
Mariniflexile sp. TRM1-10
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Mariniflexile; unclassified Mariniflexile
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
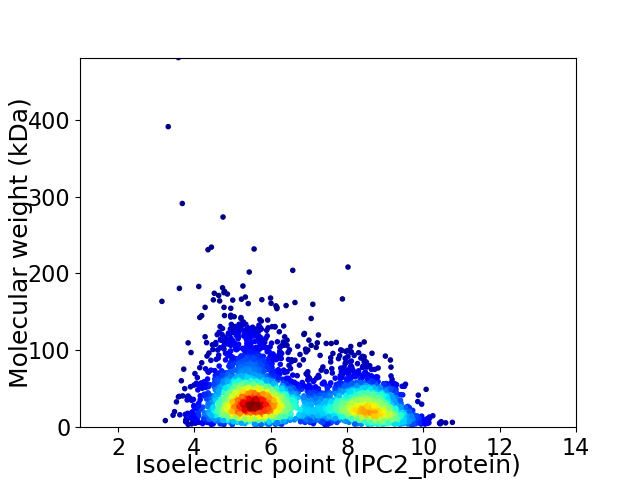
Virtual 2D-PAGE plot for 4002 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A385BTU7|A0A385BTU7_9FLAO Penicillin-binding protein 4 OS=Mariniflexile sp. TRM1-10 OX=2027857 GN=CJ739_3516 PE=4 SV=1
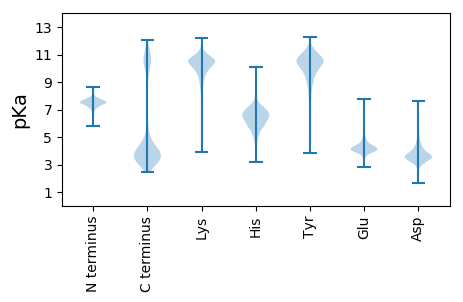
MM1 pKa = 7.3KK2 pKa = 9.74TFKK5 pKa = 10.15PLSIFTLIMLVLFAACEE22 pKa = 3.78KK23 pKa = 11.0DD24 pKa = 5.12DD25 pKa = 4.66FIEE28 pKa = 4.77TIGICPLVIATNPEE42 pKa = 3.51NDD44 pKa = 3.5ATNVPLSQVITVTFNVEE61 pKa = 4.09MNPTTITEE69 pKa = 3.78ASFIIEE75 pKa = 3.89SGAAITGTVTYY86 pKa = 10.69NGFTATFTPAAPLEE100 pKa = 4.22ISTTYY105 pKa = 9.17TGRR108 pKa = 11.84IKK110 pKa = 10.17TSVKK114 pKa = 10.42DD115 pKa = 3.77LNGNALQEE123 pKa = 4.27EE124 pKa = 5.33YY125 pKa = 11.01VWTFSTGATVSPMVISTNPEE145 pKa = 3.82DD146 pKa = 3.99NEE148 pKa = 4.12TDD150 pKa = 3.71VVINTIITATFSQQMDD166 pKa = 3.98PLTINEE172 pKa = 4.24TSFILMDD179 pKa = 4.4GSTAITGVVTYY190 pKa = 10.96NDD192 pKa = 3.15VTATFTPSANLDD204 pKa = 3.57ANTLYY209 pKa = 10.91SVTITTDD216 pKa = 2.75ATNTTGISLEE226 pKa = 4.18NNYY229 pKa = 9.35EE230 pKa = 3.86WSFTTGTLISPMVISTDD247 pKa = 3.6PEE249 pKa = 4.3DD250 pKa = 4.32NEE252 pKa = 4.72IDD254 pKa = 3.68VALDD258 pKa = 3.23KK259 pKa = 11.03TITATFSEE267 pKa = 4.52PMDD270 pKa = 4.18LATIDD275 pKa = 3.65EE276 pKa = 4.76TTFTLQEE283 pKa = 3.92GTNFITGTISYY294 pKa = 9.46NALTASFVPSVDD306 pKa = 3.95LLPGTLYY313 pKa = 10.86KK314 pKa = 10.27ATITTGATNVAGIPLANNYY333 pKa = 7.44EE334 pKa = 4.27WEE336 pKa = 4.37FTTAGTPFEE345 pKa = 4.1PTIDD349 pKa = 3.55LGSVEE354 pKa = 4.34RR355 pKa = 11.84FGIISGVAVSNNAGPSEE372 pKa = 3.87IHH374 pKa = 7.06DD375 pKa = 4.69LDD377 pKa = 4.27IGIYY381 pKa = 9.28PGARR385 pKa = 11.84SSITGFFDD393 pKa = 3.29VDD395 pKa = 3.95GGPGLIFNGDD405 pKa = 3.57FYY407 pKa = 11.52AADD410 pKa = 4.25DD411 pKa = 4.65ADD413 pKa = 4.51PVPAMLLQAKK423 pKa = 9.97DD424 pKa = 4.19DD425 pKa = 3.73LTAAYY430 pKa = 9.36LAAEE434 pKa = 4.97GATSPAPATVAGDD447 pKa = 3.93LGGQTLPPGIYY458 pKa = 9.8KK459 pKa = 10.71SNSTLMIQNGNLTLDD474 pKa = 3.85GQGDD478 pKa = 4.11PNAEE482 pKa = 4.25WIFQIASDD490 pKa = 4.16FTTVGGSPYY499 pKa = 9.89PSPAGGNVILIGGAQAKK516 pKa = 9.95NIIWQVGSSAVIGDD530 pKa = 3.59YY531 pKa = 11.03TSFKK535 pKa = 11.07GNVLALTSVTMNAYY549 pKa = 8.54SQAVGRR555 pKa = 11.84MLCSNGAVTLTSTNFIYY572 pKa = 10.6RR573 pKa = 11.84PP574 pKa = 3.49
MM1 pKa = 7.3KK2 pKa = 9.74TFKK5 pKa = 10.15PLSIFTLIMLVLFAACEE22 pKa = 3.78KK23 pKa = 11.0DD24 pKa = 5.12DD25 pKa = 4.66FIEE28 pKa = 4.77TIGICPLVIATNPEE42 pKa = 3.51NDD44 pKa = 3.5ATNVPLSQVITVTFNVEE61 pKa = 4.09MNPTTITEE69 pKa = 3.78ASFIIEE75 pKa = 3.89SGAAITGTVTYY86 pKa = 10.69NGFTATFTPAAPLEE100 pKa = 4.22ISTTYY105 pKa = 9.17TGRR108 pKa = 11.84IKK110 pKa = 10.17TSVKK114 pKa = 10.42DD115 pKa = 3.77LNGNALQEE123 pKa = 4.27EE124 pKa = 5.33YY125 pKa = 11.01VWTFSTGATVSPMVISTNPEE145 pKa = 3.82DD146 pKa = 3.99NEE148 pKa = 4.12TDD150 pKa = 3.71VVINTIITATFSQQMDD166 pKa = 3.98PLTINEE172 pKa = 4.24TSFILMDD179 pKa = 4.4GSTAITGVVTYY190 pKa = 10.96NDD192 pKa = 3.15VTATFTPSANLDD204 pKa = 3.57ANTLYY209 pKa = 10.91SVTITTDD216 pKa = 2.75ATNTTGISLEE226 pKa = 4.18NNYY229 pKa = 9.35EE230 pKa = 3.86WSFTTGTLISPMVISTDD247 pKa = 3.6PEE249 pKa = 4.3DD250 pKa = 4.32NEE252 pKa = 4.72IDD254 pKa = 3.68VALDD258 pKa = 3.23KK259 pKa = 11.03TITATFSEE267 pKa = 4.52PMDD270 pKa = 4.18LATIDD275 pKa = 3.65EE276 pKa = 4.76TTFTLQEE283 pKa = 3.92GTNFITGTISYY294 pKa = 9.46NALTASFVPSVDD306 pKa = 3.95LLPGTLYY313 pKa = 10.86KK314 pKa = 10.27ATITTGATNVAGIPLANNYY333 pKa = 7.44EE334 pKa = 4.27WEE336 pKa = 4.37FTTAGTPFEE345 pKa = 4.1PTIDD349 pKa = 3.55LGSVEE354 pKa = 4.34RR355 pKa = 11.84FGIISGVAVSNNAGPSEE372 pKa = 3.87IHH374 pKa = 7.06DD375 pKa = 4.69LDD377 pKa = 4.27IGIYY381 pKa = 9.28PGARR385 pKa = 11.84SSITGFFDD393 pKa = 3.29VDD395 pKa = 3.95GGPGLIFNGDD405 pKa = 3.57FYY407 pKa = 11.52AADD410 pKa = 4.25DD411 pKa = 4.65ADD413 pKa = 4.51PVPAMLLQAKK423 pKa = 9.97DD424 pKa = 4.19DD425 pKa = 3.73LTAAYY430 pKa = 9.36LAAEE434 pKa = 4.97GATSPAPATVAGDD447 pKa = 3.93LGGQTLPPGIYY458 pKa = 9.8KK459 pKa = 10.71SNSTLMIQNGNLTLDD474 pKa = 3.85GQGDD478 pKa = 4.11PNAEE482 pKa = 4.25WIFQIASDD490 pKa = 4.16FTTVGGSPYY499 pKa = 9.89PSPAGGNVILIGGAQAKK516 pKa = 9.95NIIWQVGSSAVIGDD530 pKa = 3.59YY531 pKa = 11.03TSFKK535 pKa = 11.07GNVLALTSVTMNAYY549 pKa = 8.54SQAVGRR555 pKa = 11.84MLCSNGAVTLTSTNFIYY572 pKa = 10.6RR573 pKa = 11.84PP574 pKa = 3.49
Molecular weight: 60.44 kDa
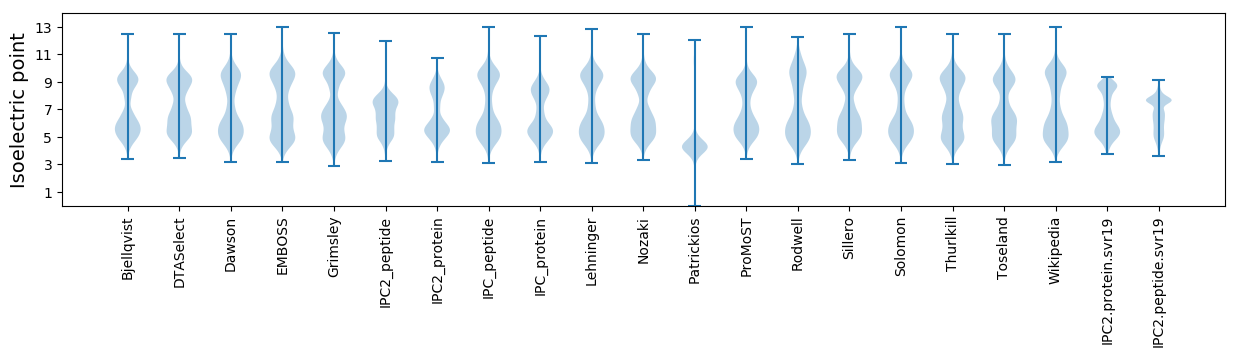
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A385BK77|A0A385BK77_9FLAO Uncharacterized protein OS=Mariniflexile sp. TRM1-10 OX=2027857 GN=CJ739_386 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.6LSVSSEE48 pKa = 4.04SRR50 pKa = 11.84HH51 pKa = 5.63KK52 pKa = 10.69KK53 pKa = 9.55
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.6LSVSSEE48 pKa = 4.04SRR50 pKa = 11.84HH51 pKa = 5.63KK52 pKa = 10.69KK53 pKa = 9.55
Molecular weight: 6.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1401172 |
29 |
4580 |
350.1 |
39.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.167 ± 0.039 | 0.767 ± 0.016 |
5.611 ± 0.028 | 6.336 ± 0.041 |
5.2 ± 0.031 | 6.363 ± 0.038 |
1.831 ± 0.016 | 8.086 ± 0.038 |
7.836 ± 0.05 | 9.107 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.117 ± 0.019 | 6.584 ± 0.038 |
3.384 ± 0.024 | 3.262 ± 0.019 |
3.205 ± 0.024 | 6.58 ± 0.031 |
6.073 ± 0.052 | 6.086 ± 0.032 |
1.132 ± 0.016 | 4.273 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
