
delta proteobacterium MLMS-1
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; unclassified Deltaproteobacteria
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
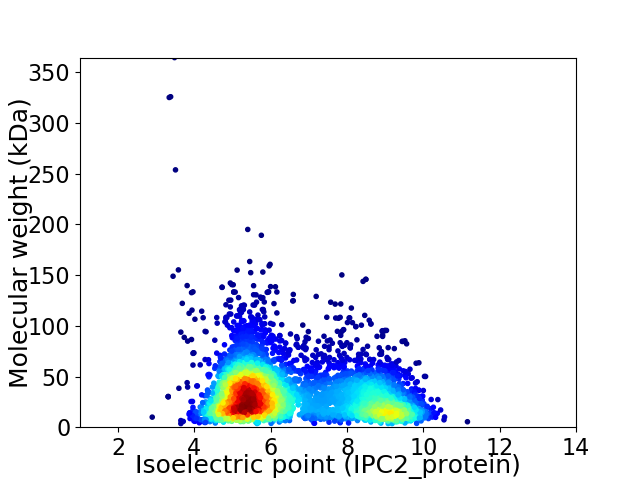
Virtual 2D-PAGE plot for 4800 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q1NSR5|Q1NSR5_9DELT Uncharacterized protein OS=delta proteobacterium MLMS-1 OX=262489 GN=MldDRAFT_1382 PE=4 SV=1
MM1 pKa = 6.6YY2 pKa = 10.16QSRR5 pKa = 11.84HH6 pKa = 4.83PQSFAVLLVAFIYY19 pKa = 10.55LFTAAFWVPPVAAAEE34 pKa = 4.01ATEE37 pKa = 3.93AAKK40 pKa = 10.2IVALRR45 pKa = 11.84GSAVAIDD52 pKa = 3.66ADD54 pKa = 4.15GVEE57 pKa = 4.43RR58 pKa = 11.84TLAPGDD64 pKa = 3.86SLAVSDD70 pKa = 5.02TIRR73 pKa = 11.84TGPRR77 pKa = 11.84GRR79 pKa = 11.84LQLMFSDD86 pKa = 3.69NTIISLGVNAEE97 pKa = 4.33LEE99 pKa = 4.06ISSYY103 pKa = 11.14HH104 pKa = 7.3LDD106 pKa = 3.58EE107 pKa = 6.29DD108 pKa = 3.94SGEE111 pKa = 4.21LATTVNRR118 pKa = 11.84GAFRR122 pKa = 11.84VMGGSIARR130 pKa = 11.84TSPEE134 pKa = 4.05SFTTDD139 pKa = 3.1TPSATIGIRR148 pKa = 11.84GSMYY152 pKa = 10.52AGRR155 pKa = 11.84VSNGALTVVFQGGVGITVSNPAGTVEE181 pKa = 3.81ITTPGMGTRR190 pKa = 11.84VRR192 pKa = 11.84SRR194 pKa = 11.84DD195 pKa = 3.5EE196 pKa = 4.17APEE199 pKa = 3.85EE200 pKa = 3.82PRR202 pKa = 11.84PFGEE206 pKa = 5.4DD207 pKa = 3.04DD208 pKa = 3.59LAEE211 pKa = 4.62LEE213 pKa = 4.58DD214 pKa = 3.97EE215 pKa = 4.5EE216 pKa = 4.67EE217 pKa = 4.04ATEE220 pKa = 4.14EE221 pKa = 4.05EE222 pKa = 4.47EE223 pKa = 4.29EE224 pKa = 4.21ATEE227 pKa = 4.45EE228 pKa = 4.16EE229 pKa = 4.23ATEE232 pKa = 4.2DD233 pKa = 3.91DD234 pKa = 4.15EE235 pKa = 5.52AAEE238 pKa = 4.05EE239 pKa = 4.18QPADD243 pKa = 3.62EE244 pKa = 4.84EE245 pKa = 4.43EE246 pKa = 4.24WDD248 pKa = 4.03EE249 pKa = 4.19EE250 pKa = 4.31ALDD253 pKa = 4.16EE254 pKa = 5.8ADD256 pKa = 3.45TTTDD260 pKa = 3.11TTEE263 pKa = 4.0TTTTTEE269 pKa = 4.17VEE271 pKa = 4.46DD272 pKa = 4.01VATEE276 pKa = 4.36ASQDD280 pKa = 3.65EE281 pKa = 4.38QQDD284 pKa = 3.78DD285 pKa = 4.04VTQKK289 pKa = 11.09AEE291 pKa = 4.01EE292 pKa = 4.15TATEE296 pKa = 3.83EE297 pKa = 4.49DD298 pKa = 3.61ATHH301 pKa = 6.65SGRR304 pKa = 11.84AIWVWQEE311 pKa = 3.64EE312 pKa = 4.75GEE314 pKa = 4.19EE315 pKa = 4.52AEE317 pKa = 4.22VQLISFEE324 pKa = 4.52DD325 pKa = 3.37HH326 pKa = 6.23WSEE329 pKa = 5.23HH330 pKa = 7.21DD331 pKa = 5.02DD332 pKa = 4.32GHH334 pKa = 7.72DD335 pKa = 4.29DD336 pKa = 4.7LPKK339 pKa = 9.91DD340 pKa = 4.03TYY342 pKa = 10.74TGPHH346 pKa = 6.23WDD348 pKa = 3.93EE349 pKa = 4.82ASGDD353 pKa = 3.77DD354 pKa = 3.2WWWRR358 pKa = 11.84GGYY361 pKa = 8.02EE362 pKa = 4.18DD363 pKa = 5.33RR364 pKa = 11.84HH365 pKa = 5.32EE366 pKa = 4.58FWVKK370 pKa = 8.72MDD372 pKa = 3.78NYY374 pKa = 10.43DD375 pKa = 3.32GHH377 pKa = 7.66SEE379 pKa = 4.67GNLQDD384 pKa = 3.09SWFSFYY390 pKa = 11.29GLEE393 pKa = 4.23TTTGQMPGSGGSQYY407 pKa = 11.29SGYY410 pKa = 10.98FMGADD415 pKa = 3.89HH416 pKa = 7.53DD417 pKa = 4.35PSYY420 pKa = 10.63PNPDD424 pKa = 3.08FGEE427 pKa = 4.33FQFVTNWRR435 pKa = 11.84TGKK438 pKa = 10.18AVGYY442 pKa = 9.91FEE444 pKa = 5.79EE445 pKa = 6.85GDD447 pKa = 3.48DD448 pKa = 3.53TRR450 pKa = 11.84VFFHH454 pKa = 7.38ADD456 pKa = 2.99VSGTTLSNVQIMGTHH471 pKa = 6.08YY472 pKa = 10.96YY473 pKa = 10.83DD474 pKa = 4.72DD475 pKa = 4.22GEE477 pKa = 4.02EE478 pKa = 4.17RR479 pKa = 11.84YY480 pKa = 9.97EE481 pKa = 4.09YY482 pKa = 11.12VAGDD486 pKa = 3.65SNFFSFYY493 pKa = 9.66GAYY496 pKa = 9.42YY497 pKa = 10.04QGFGMTTQGEE507 pKa = 4.55NYY509 pKa = 9.82SLEE512 pKa = 4.04NMEE515 pKa = 6.24SVVQWSAISAGFRR528 pKa = 11.84EE529 pKa = 3.99TDD531 pKa = 3.22DD532 pKa = 3.56NYY534 pKa = 11.47SYY536 pKa = 11.87VDD538 pKa = 3.51NTATSHH544 pKa = 4.56WQGFVTGAAINVAGTDD560 pKa = 3.79DD561 pKa = 4.16NNLLFMNDD569 pKa = 3.59DD570 pKa = 3.69PEE572 pKa = 5.18AFNFLINRR580 pKa = 11.84EE581 pKa = 3.82QGMFSGEE588 pKa = 3.9INISNIFTDD597 pKa = 3.1TDD599 pKa = 3.22GDD601 pKa = 4.12YY602 pKa = 11.17EE603 pKa = 4.86LNLGAQGTYY612 pKa = 10.24VADD615 pKa = 3.58NFMLGNIDD623 pKa = 3.64GTVTNHH629 pKa = 5.97DD630 pKa = 4.1TEE632 pKa = 4.72TSGSLNQYY640 pKa = 10.4GNMIVASPEE649 pKa = 3.43ISMDD653 pKa = 3.32EE654 pKa = 4.4HH655 pKa = 8.3ISWGYY660 pKa = 9.32WSVSFDD666 pKa = 4.38DD667 pKa = 3.96PHH669 pKa = 6.46STDD672 pKa = 2.51TTYY675 pKa = 11.82ALDD678 pKa = 3.66PRR680 pKa = 11.84ASFWVAGEE688 pKa = 4.21STPAEE693 pKa = 4.21YY694 pKa = 11.04VEE696 pKa = 4.64GLIDD700 pKa = 3.69NDD702 pKa = 3.86YY703 pKa = 10.54VGQYY707 pKa = 9.52TGGAQGIKK715 pKa = 10.29VEE717 pKa = 4.19NDD719 pKa = 3.05VIAALTNGVTDD730 pKa = 3.34LTIYY734 pKa = 10.5FGSGTMAGSISFDD747 pKa = 3.25EE748 pKa = 4.31VSLNVDD754 pKa = 3.42SGSVSSSGFSADD766 pKa = 3.43FSNMNSSGLKK776 pKa = 10.16GGFFGDD782 pKa = 3.6QAQSIGGNFQAEE794 pKa = 4.66DD795 pKa = 3.58GDD797 pKa = 4.07ADD799 pKa = 3.73YY800 pKa = 11.63SGVFGADD807 pKa = 2.59MEE809 pKa = 4.55MPHH812 pKa = 6.58
MM1 pKa = 6.6YY2 pKa = 10.16QSRR5 pKa = 11.84HH6 pKa = 4.83PQSFAVLLVAFIYY19 pKa = 10.55LFTAAFWVPPVAAAEE34 pKa = 4.01ATEE37 pKa = 3.93AAKK40 pKa = 10.2IVALRR45 pKa = 11.84GSAVAIDD52 pKa = 3.66ADD54 pKa = 4.15GVEE57 pKa = 4.43RR58 pKa = 11.84TLAPGDD64 pKa = 3.86SLAVSDD70 pKa = 5.02TIRR73 pKa = 11.84TGPRR77 pKa = 11.84GRR79 pKa = 11.84LQLMFSDD86 pKa = 3.69NTIISLGVNAEE97 pKa = 4.33LEE99 pKa = 4.06ISSYY103 pKa = 11.14HH104 pKa = 7.3LDD106 pKa = 3.58EE107 pKa = 6.29DD108 pKa = 3.94SGEE111 pKa = 4.21LATTVNRR118 pKa = 11.84GAFRR122 pKa = 11.84VMGGSIARR130 pKa = 11.84TSPEE134 pKa = 4.05SFTTDD139 pKa = 3.1TPSATIGIRR148 pKa = 11.84GSMYY152 pKa = 10.52AGRR155 pKa = 11.84VSNGALTVVFQGGVGITVSNPAGTVEE181 pKa = 3.81ITTPGMGTRR190 pKa = 11.84VRR192 pKa = 11.84SRR194 pKa = 11.84DD195 pKa = 3.5EE196 pKa = 4.17APEE199 pKa = 3.85EE200 pKa = 3.82PRR202 pKa = 11.84PFGEE206 pKa = 5.4DD207 pKa = 3.04DD208 pKa = 3.59LAEE211 pKa = 4.62LEE213 pKa = 4.58DD214 pKa = 3.97EE215 pKa = 4.5EE216 pKa = 4.67EE217 pKa = 4.04ATEE220 pKa = 4.14EE221 pKa = 4.05EE222 pKa = 4.47EE223 pKa = 4.29EE224 pKa = 4.21ATEE227 pKa = 4.45EE228 pKa = 4.16EE229 pKa = 4.23ATEE232 pKa = 4.2DD233 pKa = 3.91DD234 pKa = 4.15EE235 pKa = 5.52AAEE238 pKa = 4.05EE239 pKa = 4.18QPADD243 pKa = 3.62EE244 pKa = 4.84EE245 pKa = 4.43EE246 pKa = 4.24WDD248 pKa = 4.03EE249 pKa = 4.19EE250 pKa = 4.31ALDD253 pKa = 4.16EE254 pKa = 5.8ADD256 pKa = 3.45TTTDD260 pKa = 3.11TTEE263 pKa = 4.0TTTTTEE269 pKa = 4.17VEE271 pKa = 4.46DD272 pKa = 4.01VATEE276 pKa = 4.36ASQDD280 pKa = 3.65EE281 pKa = 4.38QQDD284 pKa = 3.78DD285 pKa = 4.04VTQKK289 pKa = 11.09AEE291 pKa = 4.01EE292 pKa = 4.15TATEE296 pKa = 3.83EE297 pKa = 4.49DD298 pKa = 3.61ATHH301 pKa = 6.65SGRR304 pKa = 11.84AIWVWQEE311 pKa = 3.64EE312 pKa = 4.75GEE314 pKa = 4.19EE315 pKa = 4.52AEE317 pKa = 4.22VQLISFEE324 pKa = 4.52DD325 pKa = 3.37HH326 pKa = 6.23WSEE329 pKa = 5.23HH330 pKa = 7.21DD331 pKa = 5.02DD332 pKa = 4.32GHH334 pKa = 7.72DD335 pKa = 4.29DD336 pKa = 4.7LPKK339 pKa = 9.91DD340 pKa = 4.03TYY342 pKa = 10.74TGPHH346 pKa = 6.23WDD348 pKa = 3.93EE349 pKa = 4.82ASGDD353 pKa = 3.77DD354 pKa = 3.2WWWRR358 pKa = 11.84GGYY361 pKa = 8.02EE362 pKa = 4.18DD363 pKa = 5.33RR364 pKa = 11.84HH365 pKa = 5.32EE366 pKa = 4.58FWVKK370 pKa = 8.72MDD372 pKa = 3.78NYY374 pKa = 10.43DD375 pKa = 3.32GHH377 pKa = 7.66SEE379 pKa = 4.67GNLQDD384 pKa = 3.09SWFSFYY390 pKa = 11.29GLEE393 pKa = 4.23TTTGQMPGSGGSQYY407 pKa = 11.29SGYY410 pKa = 10.98FMGADD415 pKa = 3.89HH416 pKa = 7.53DD417 pKa = 4.35PSYY420 pKa = 10.63PNPDD424 pKa = 3.08FGEE427 pKa = 4.33FQFVTNWRR435 pKa = 11.84TGKK438 pKa = 10.18AVGYY442 pKa = 9.91FEE444 pKa = 5.79EE445 pKa = 6.85GDD447 pKa = 3.48DD448 pKa = 3.53TRR450 pKa = 11.84VFFHH454 pKa = 7.38ADD456 pKa = 2.99VSGTTLSNVQIMGTHH471 pKa = 6.08YY472 pKa = 10.96YY473 pKa = 10.83DD474 pKa = 4.72DD475 pKa = 4.22GEE477 pKa = 4.02EE478 pKa = 4.17RR479 pKa = 11.84YY480 pKa = 9.97EE481 pKa = 4.09YY482 pKa = 11.12VAGDD486 pKa = 3.65SNFFSFYY493 pKa = 9.66GAYY496 pKa = 9.42YY497 pKa = 10.04QGFGMTTQGEE507 pKa = 4.55NYY509 pKa = 9.82SLEE512 pKa = 4.04NMEE515 pKa = 6.24SVVQWSAISAGFRR528 pKa = 11.84EE529 pKa = 3.99TDD531 pKa = 3.22DD532 pKa = 3.56NYY534 pKa = 11.47SYY536 pKa = 11.87VDD538 pKa = 3.51NTATSHH544 pKa = 4.56WQGFVTGAAINVAGTDD560 pKa = 3.79DD561 pKa = 4.16NNLLFMNDD569 pKa = 3.59DD570 pKa = 3.69PEE572 pKa = 5.18AFNFLINRR580 pKa = 11.84EE581 pKa = 3.82QGMFSGEE588 pKa = 3.9INISNIFTDD597 pKa = 3.1TDD599 pKa = 3.22GDD601 pKa = 4.12YY602 pKa = 11.17EE603 pKa = 4.86LNLGAQGTYY612 pKa = 10.24VADD615 pKa = 3.58NFMLGNIDD623 pKa = 3.64GTVTNHH629 pKa = 5.97DD630 pKa = 4.1TEE632 pKa = 4.72TSGSLNQYY640 pKa = 10.4GNMIVASPEE649 pKa = 3.43ISMDD653 pKa = 3.32EE654 pKa = 4.4HH655 pKa = 8.3ISWGYY660 pKa = 9.32WSVSFDD666 pKa = 4.38DD667 pKa = 3.96PHH669 pKa = 6.46STDD672 pKa = 2.51TTYY675 pKa = 11.82ALDD678 pKa = 3.66PRR680 pKa = 11.84ASFWVAGEE688 pKa = 4.21STPAEE693 pKa = 4.21YY694 pKa = 11.04VEE696 pKa = 4.64GLIDD700 pKa = 3.69NDD702 pKa = 3.86YY703 pKa = 10.54VGQYY707 pKa = 9.52TGGAQGIKK715 pKa = 10.29VEE717 pKa = 4.19NDD719 pKa = 3.05VIAALTNGVTDD730 pKa = 3.34LTIYY734 pKa = 10.5FGSGTMAGSISFDD747 pKa = 3.25EE748 pKa = 4.31VSLNVDD754 pKa = 3.42SGSVSSSGFSADD766 pKa = 3.43FSNMNSSGLKK776 pKa = 10.16GGFFGDD782 pKa = 3.6QAQSIGGNFQAEE794 pKa = 4.66DD795 pKa = 3.58GDD797 pKa = 4.07ADD799 pKa = 3.73YY800 pKa = 11.63SGVFGADD807 pKa = 2.59MEE809 pKa = 4.55MPHH812 pKa = 6.58
Molecular weight: 88.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q1NM73|Q1NM73_9DELT Pseudouridine synthase OS=delta proteobacterium MLMS-1 OX=262489 GN=MldDRAFT_3455 PE=3 SV=1
MM1 pKa = 7.74SKK3 pKa = 8.95RR4 pKa = 11.84TYY6 pKa = 10.22QPSNTKK12 pKa = 8.42RR13 pKa = 11.84HH14 pKa = 5.05RR15 pKa = 11.84THH17 pKa = 6.71GFRR20 pKa = 11.84VRR22 pKa = 11.84MSTKK26 pKa = 10.09NGRR29 pKa = 11.84AVINRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 8.8RR42 pKa = 11.84LAVV45 pKa = 3.5
MM1 pKa = 7.74SKK3 pKa = 8.95RR4 pKa = 11.84TYY6 pKa = 10.22QPSNTKK12 pKa = 8.42RR13 pKa = 11.84HH14 pKa = 5.05RR15 pKa = 11.84THH17 pKa = 6.71GFRR20 pKa = 11.84VRR22 pKa = 11.84MSTKK26 pKa = 10.09NGRR29 pKa = 11.84AVINRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 8.8RR42 pKa = 11.84LAVV45 pKa = 3.5
Molecular weight: 5.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1571437 |
32 |
3532 |
327.4 |
36.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.751 ± 0.05 | 1.14 ± 0.016 |
5.269 ± 0.031 | 6.919 ± 0.038 |
3.589 ± 0.019 | 7.92 ± 0.034 |
2.306 ± 0.016 | 5.128 ± 0.028 |
3.619 ± 0.028 | 11.601 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.154 ± 0.016 | 2.897 ± 0.024 |
5.186 ± 0.032 | 4.274 ± 0.027 |
7.4 ± 0.042 | 4.91 ± 0.024 |
4.688 ± 0.039 | 6.553 ± 0.027 |
1.155 ± 0.014 | 2.538 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
