
Nocardioides seonyuensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides
Average proteome isoelectric point is 5.93
Get precalculated fractions of proteins
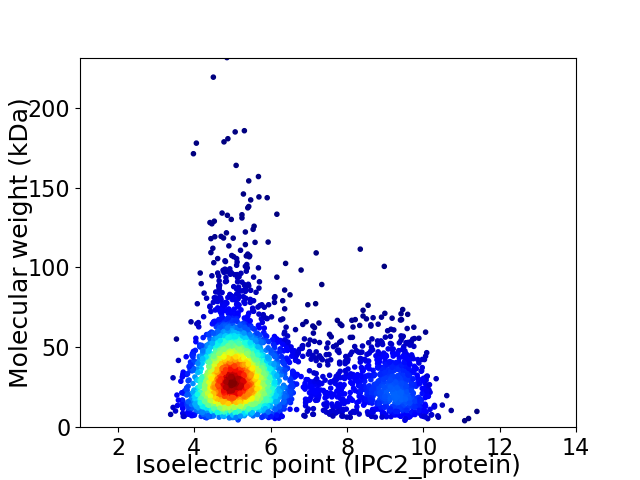
Virtual 2D-PAGE plot for 3712 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1BLV0|A0A4V1BLV0_9ACTN Uncharacterized protein OS=Nocardioides seonyuensis OX=2518371 GN=EXE58_00740 PE=4 SV=1
MM1 pKa = 8.03RR2 pKa = 11.84LGTSAAGVAALALILSACGGADD24 pKa = 3.1AGEE27 pKa = 4.45SGSGEE32 pKa = 4.17GAGGTVAVDD41 pKa = 4.1GSSTVEE47 pKa = 4.04PMSKK51 pKa = 10.12AASEE55 pKa = 4.15LLSEE59 pKa = 4.4EE60 pKa = 4.14NADD63 pKa = 3.62VRR65 pKa = 11.84VTVAASGTGGGFEE78 pKa = 4.26KK79 pKa = 10.75FCVGEE84 pKa = 4.1SDD86 pKa = 3.75ISDD89 pKa = 3.29ASRR92 pKa = 11.84PIKK95 pKa = 10.17EE96 pKa = 4.25DD97 pKa = 3.31EE98 pKa = 4.12EE99 pKa = 5.21VPVCEE104 pKa = 4.42EE105 pKa = 3.75NGVEE109 pKa = 4.11YY110 pKa = 10.84TEE112 pKa = 4.6LQVATDD118 pKa = 3.41ALTVVVHH125 pKa = 6.89PDD127 pKa = 3.32LAVDD131 pKa = 4.46CLTTEE136 pKa = 3.92QLVEE140 pKa = 4.11LWHH143 pKa = 6.14PTSKK147 pKa = 8.69VTNWNEE153 pKa = 3.81LDD155 pKa = 3.67PSFPDD160 pKa = 3.54QEE162 pKa = 4.12IALFGPGTDD171 pKa = 3.19SGTYY175 pKa = 10.12DD176 pKa = 3.4YY177 pKa = 10.61MAADD181 pKa = 4.11VIGDD185 pKa = 3.58EE186 pKa = 4.71SEE188 pKa = 4.36TTRR191 pKa = 11.84SDD193 pKa = 3.83YY194 pKa = 11.09EE195 pKa = 4.1SSEE198 pKa = 4.12DD199 pKa = 4.99DD200 pKa = 3.42NVLVQGVSGTEE211 pKa = 3.72GATGYY216 pKa = 10.79FGFTYY221 pKa = 10.65YY222 pKa = 10.73EE223 pKa = 4.16EE224 pKa = 4.59NADD227 pKa = 3.88SLKK230 pKa = 11.08ALAIDD235 pKa = 4.67DD236 pKa = 4.64GNGCVEE242 pKa = 4.41PSVEE246 pKa = 3.93TAQAGEE252 pKa = 4.27YY253 pKa = 9.36TPLARR258 pKa = 11.84PLFIYY263 pKa = 10.4VSNASYY269 pKa = 11.21AEE271 pKa = 4.12KK272 pKa = 10.0PAVAEE277 pKa = 3.96YY278 pKa = 10.98VDD280 pKa = 5.86FYY282 pKa = 11.16IEE284 pKa = 4.01NLEE287 pKa = 4.38QIATAAKK294 pKa = 9.29FIPLSEE300 pKa = 4.11EE301 pKa = 4.09LYY303 pKa = 10.89EE304 pKa = 4.2EE305 pKa = 4.31TKK307 pKa = 10.76SALEE311 pKa = 4.93GISSS315 pKa = 3.58
MM1 pKa = 8.03RR2 pKa = 11.84LGTSAAGVAALALILSACGGADD24 pKa = 3.1AGEE27 pKa = 4.45SGSGEE32 pKa = 4.17GAGGTVAVDD41 pKa = 4.1GSSTVEE47 pKa = 4.04PMSKK51 pKa = 10.12AASEE55 pKa = 4.15LLSEE59 pKa = 4.4EE60 pKa = 4.14NADD63 pKa = 3.62VRR65 pKa = 11.84VTVAASGTGGGFEE78 pKa = 4.26KK79 pKa = 10.75FCVGEE84 pKa = 4.1SDD86 pKa = 3.75ISDD89 pKa = 3.29ASRR92 pKa = 11.84PIKK95 pKa = 10.17EE96 pKa = 4.25DD97 pKa = 3.31EE98 pKa = 4.12EE99 pKa = 5.21VPVCEE104 pKa = 4.42EE105 pKa = 3.75NGVEE109 pKa = 4.11YY110 pKa = 10.84TEE112 pKa = 4.6LQVATDD118 pKa = 3.41ALTVVVHH125 pKa = 6.89PDD127 pKa = 3.32LAVDD131 pKa = 4.46CLTTEE136 pKa = 3.92QLVEE140 pKa = 4.11LWHH143 pKa = 6.14PTSKK147 pKa = 8.69VTNWNEE153 pKa = 3.81LDD155 pKa = 3.67PSFPDD160 pKa = 3.54QEE162 pKa = 4.12IALFGPGTDD171 pKa = 3.19SGTYY175 pKa = 10.12DD176 pKa = 3.4YY177 pKa = 10.61MAADD181 pKa = 4.11VIGDD185 pKa = 3.58EE186 pKa = 4.71SEE188 pKa = 4.36TTRR191 pKa = 11.84SDD193 pKa = 3.83YY194 pKa = 11.09EE195 pKa = 4.1SSEE198 pKa = 4.12DD199 pKa = 4.99DD200 pKa = 3.42NVLVQGVSGTEE211 pKa = 3.72GATGYY216 pKa = 10.79FGFTYY221 pKa = 10.65YY222 pKa = 10.73EE223 pKa = 4.16EE224 pKa = 4.59NADD227 pKa = 3.88SLKK230 pKa = 11.08ALAIDD235 pKa = 4.67DD236 pKa = 4.64GNGCVEE242 pKa = 4.41PSVEE246 pKa = 3.93TAQAGEE252 pKa = 4.27YY253 pKa = 9.36TPLARR258 pKa = 11.84PLFIYY263 pKa = 10.4VSNASYY269 pKa = 11.21AEE271 pKa = 4.12KK272 pKa = 10.0PAVAEE277 pKa = 3.96YY278 pKa = 10.98VDD280 pKa = 5.86FYY282 pKa = 11.16IEE284 pKa = 4.01NLEE287 pKa = 4.38QIATAAKK294 pKa = 9.29FIPLSEE300 pKa = 4.11EE301 pKa = 4.09LYY303 pKa = 10.89EE304 pKa = 4.2EE305 pKa = 4.31TKK307 pKa = 10.76SALEE311 pKa = 4.93GISSS315 pKa = 3.58
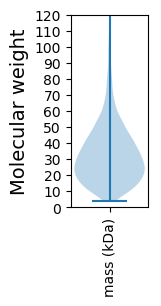
Molecular weight: 33.04 kDa
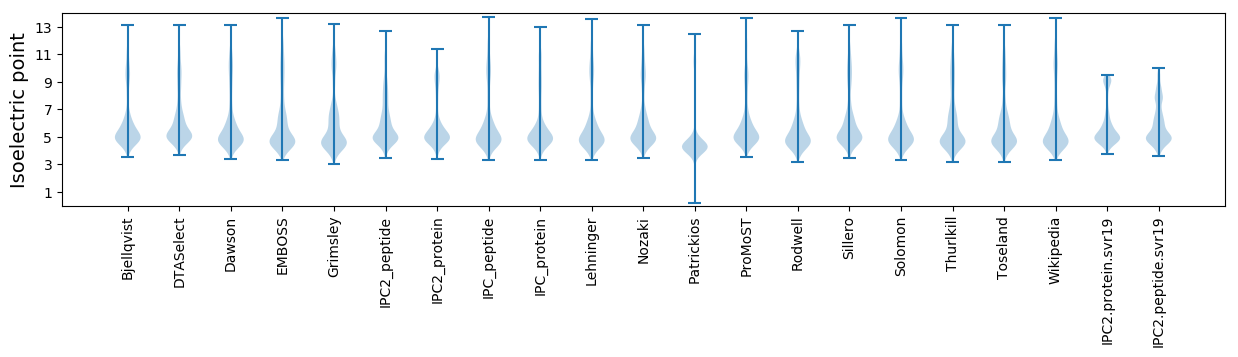
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P7IJ59|A0A4P7IJ59_9ACTN Urease accessory protein UreG OS=Nocardioides seonyuensis OX=2518371 GN=ureG PE=3 SV=1
MM1 pKa = 7.46LRR3 pKa = 11.84RR4 pKa = 11.84SRR6 pKa = 11.84ARR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84PRR14 pKa = 11.84PLGSRR19 pKa = 11.84PPRR22 pKa = 11.84PRR24 pKa = 11.84PRR26 pKa = 11.84ARR28 pKa = 11.84RR29 pKa = 11.84PLRR32 pKa = 11.84VRR34 pKa = 11.84RR35 pKa = 11.84TRR37 pKa = 11.84LLPRR41 pKa = 11.84PCHH44 pKa = 5.54RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84SPGRR53 pKa = 11.84PSAAPGRR60 pKa = 11.84LLATPGRR67 pKa = 11.84AGPQSGRR74 pKa = 11.84RR75 pKa = 11.84TTSRR79 pKa = 11.84AGSPVRR85 pKa = 4.0
MM1 pKa = 7.46LRR3 pKa = 11.84RR4 pKa = 11.84SRR6 pKa = 11.84ARR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84PRR14 pKa = 11.84PLGSRR19 pKa = 11.84PPRR22 pKa = 11.84PRR24 pKa = 11.84PRR26 pKa = 11.84ARR28 pKa = 11.84RR29 pKa = 11.84PLRR32 pKa = 11.84VRR34 pKa = 11.84RR35 pKa = 11.84TRR37 pKa = 11.84LLPRR41 pKa = 11.84PCHH44 pKa = 5.54RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84SPGRR53 pKa = 11.84PSAAPGRR60 pKa = 11.84LLATPGRR67 pKa = 11.84AGPQSGRR74 pKa = 11.84RR75 pKa = 11.84TTSRR79 pKa = 11.84AGSPVRR85 pKa = 4.0
Molecular weight: 9.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1194092 |
33 |
2236 |
321.7 |
34.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.682 ± 0.052 | 0.739 ± 0.011 |
6.507 ± 0.032 | 6.033 ± 0.038 |
2.785 ± 0.021 | 9.107 ± 0.04 |
2.299 ± 0.022 | 3.393 ± 0.022 |
1.985 ± 0.028 | 10.285 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.941 ± 0.014 | 1.714 ± 0.02 |
5.573 ± 0.032 | 2.764 ± 0.018 |
7.689 ± 0.045 | 5.565 ± 0.029 |
6.069 ± 0.037 | 9.487 ± 0.043 |
1.574 ± 0.016 | 1.809 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
