
Kotonkan virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Ephemerovirus; Kotonkan ephemerovirus
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
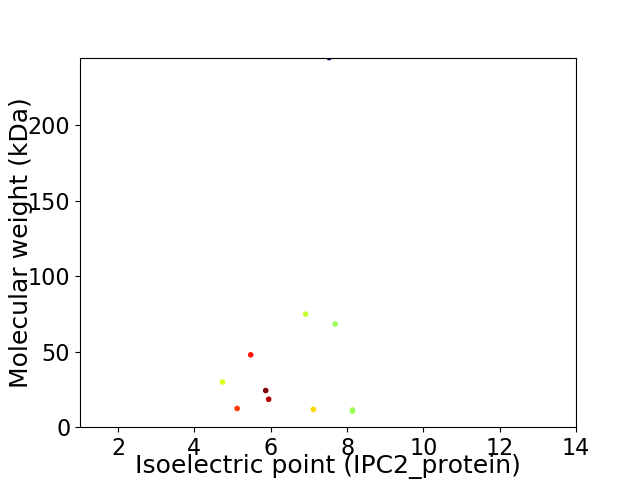
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H8XWF2|H8XWF2_9RHAB Matrix protein OS=Kotonkan virus OX=318836 GN=M PE=4 SV=1
MM1 pKa = 7.59EE2 pKa = 5.86PYY4 pKa = 10.38KK5 pKa = 10.72LSEE8 pKa = 4.17KK9 pKa = 8.22MTGYY13 pKa = 10.99DD14 pKa = 3.44LVKK17 pKa = 10.51LRR19 pKa = 11.84EE20 pKa = 4.09VLNEE24 pKa = 3.87VDD26 pKa = 6.22DD27 pKa = 4.57EE28 pKa = 4.75PEE30 pKa = 3.89DD31 pKa = 3.82TKK33 pKa = 11.32VDD35 pKa = 4.49DD36 pKa = 4.24VTQSPNPLFEE46 pKa = 4.23TSMKK50 pKa = 10.92GNDD53 pKa = 3.53SDD55 pKa = 4.85EE56 pKa = 4.15EE57 pKa = 4.02WGEE60 pKa = 4.11IISNMSNNVCSVNNIINKK78 pKa = 9.84KK79 pKa = 9.55SDD81 pKa = 3.01NDD83 pKa = 3.66APEE86 pKa = 4.07GRR88 pKa = 11.84NDD90 pKa = 3.62VIKK93 pKa = 10.14KK94 pKa = 10.07GRR96 pKa = 11.84MASEE100 pKa = 4.23EE101 pKa = 4.5SNSNLPLMWDD111 pKa = 3.88NNWVMICDD119 pKa = 5.07DD120 pKa = 4.67ISCEE124 pKa = 3.81MSKK127 pKa = 10.36AHH129 pKa = 6.62SLLSLFCLKK138 pKa = 10.8EE139 pKa = 3.8NVDD142 pKa = 3.54YY143 pKa = 11.3KK144 pKa = 10.89FIVNEE149 pKa = 3.88QTLTVQKK156 pKa = 10.91LKK158 pKa = 10.91FNSDD162 pKa = 3.97DD163 pKa = 3.88NPQEE167 pKa = 4.19ASGVTYY173 pKa = 8.58KK174 pKa = 9.76TANYY178 pKa = 10.4SRR180 pKa = 11.84FDD182 pKa = 3.64EE183 pKa = 4.77PEE185 pKa = 4.0EE186 pKa = 4.26SSLKK190 pKa = 10.53YY191 pKa = 10.19KK192 pKa = 10.15IHH194 pKa = 6.04TRR196 pKa = 11.84LDD198 pKa = 3.16EE199 pKa = 4.76GIKK202 pKa = 10.19FKK204 pKa = 10.98KK205 pKa = 10.61LNGKK209 pKa = 10.09GYY211 pKa = 10.18IRR213 pKa = 11.84VSWFTDD219 pKa = 3.56GVYY222 pKa = 10.8QNILDD227 pKa = 5.13DD228 pKa = 4.3IDD230 pKa = 3.67WNNTKK235 pKa = 10.16SEE237 pKa = 4.19EE238 pKa = 4.07EE239 pKa = 3.74AVIRR243 pKa = 11.84VLKK246 pKa = 9.88SAKK249 pKa = 8.83IYY251 pKa = 11.24KK252 pKa = 9.01MIKK255 pKa = 7.81KK256 pKa = 7.74TCEE259 pKa = 3.35II260 pKa = 4.25
MM1 pKa = 7.59EE2 pKa = 5.86PYY4 pKa = 10.38KK5 pKa = 10.72LSEE8 pKa = 4.17KK9 pKa = 8.22MTGYY13 pKa = 10.99DD14 pKa = 3.44LVKK17 pKa = 10.51LRR19 pKa = 11.84EE20 pKa = 4.09VLNEE24 pKa = 3.87VDD26 pKa = 6.22DD27 pKa = 4.57EE28 pKa = 4.75PEE30 pKa = 3.89DD31 pKa = 3.82TKK33 pKa = 11.32VDD35 pKa = 4.49DD36 pKa = 4.24VTQSPNPLFEE46 pKa = 4.23TSMKK50 pKa = 10.92GNDD53 pKa = 3.53SDD55 pKa = 4.85EE56 pKa = 4.15EE57 pKa = 4.02WGEE60 pKa = 4.11IISNMSNNVCSVNNIINKK78 pKa = 9.84KK79 pKa = 9.55SDD81 pKa = 3.01NDD83 pKa = 3.66APEE86 pKa = 4.07GRR88 pKa = 11.84NDD90 pKa = 3.62VIKK93 pKa = 10.14KK94 pKa = 10.07GRR96 pKa = 11.84MASEE100 pKa = 4.23EE101 pKa = 4.5SNSNLPLMWDD111 pKa = 3.88NNWVMICDD119 pKa = 5.07DD120 pKa = 4.67ISCEE124 pKa = 3.81MSKK127 pKa = 10.36AHH129 pKa = 6.62SLLSLFCLKK138 pKa = 10.8EE139 pKa = 3.8NVDD142 pKa = 3.54YY143 pKa = 11.3KK144 pKa = 10.89FIVNEE149 pKa = 3.88QTLTVQKK156 pKa = 10.91LKK158 pKa = 10.91FNSDD162 pKa = 3.97DD163 pKa = 3.88NPQEE167 pKa = 4.19ASGVTYY173 pKa = 8.58KK174 pKa = 9.76TANYY178 pKa = 10.4SRR180 pKa = 11.84FDD182 pKa = 3.64EE183 pKa = 4.77PEE185 pKa = 4.0EE186 pKa = 4.26SSLKK190 pKa = 10.53YY191 pKa = 10.19KK192 pKa = 10.15IHH194 pKa = 6.04TRR196 pKa = 11.84LDD198 pKa = 3.16EE199 pKa = 4.76GIKK202 pKa = 10.19FKK204 pKa = 10.98KK205 pKa = 10.61LNGKK209 pKa = 10.09GYY211 pKa = 10.18IRR213 pKa = 11.84VSWFTDD219 pKa = 3.56GVYY222 pKa = 10.8QNILDD227 pKa = 5.13DD228 pKa = 4.3IDD230 pKa = 3.67WNNTKK235 pKa = 10.16SEE237 pKa = 4.19EE238 pKa = 4.07EE239 pKa = 3.74AVIRR243 pKa = 11.84VLKK246 pKa = 9.88SAKK249 pKa = 8.83IYY251 pKa = 11.24KK252 pKa = 9.01MIKK255 pKa = 7.81KK256 pKa = 7.74TCEE259 pKa = 3.35II260 pKa = 4.25
Molecular weight: 29.93 kDa
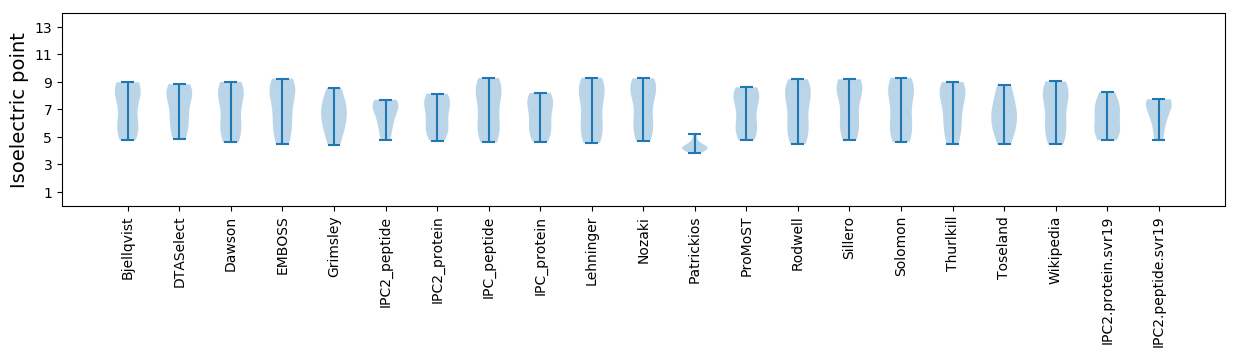
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H8XWF5|H8XWF5_9RHAB Isoform of H8XWF6 Alpha 1 protein OS=Kotonkan virus OX=318836 GN=alpha PE=4 SV=1
MM1 pKa = 7.38NKK3 pKa = 9.47VNWPSLDD10 pKa = 4.13GIKK13 pKa = 9.66LTLKK17 pKa = 9.78EE18 pKa = 4.68FPDD21 pKa = 4.55KK22 pKa = 10.98INTEE26 pKa = 3.61IRR28 pKa = 11.84EE29 pKa = 4.0FSHH32 pKa = 7.14NIVDD36 pKa = 4.52KK37 pKa = 10.88IRR39 pKa = 11.84YY40 pKa = 6.44IWVWLVVIVLLLLIIKK56 pKa = 7.72ITPLLVNLVISCKK69 pKa = 9.83RR70 pKa = 11.84CYY72 pKa = 10.47QNFITGSKK80 pKa = 9.45KK81 pKa = 10.42LKK83 pKa = 10.35EE84 pKa = 4.43EE85 pKa = 3.99EE86 pKa = 4.21TEE88 pKa = 4.06IAA90 pKa = 4.37
MM1 pKa = 7.38NKK3 pKa = 9.47VNWPSLDD10 pKa = 4.13GIKK13 pKa = 9.66LTLKK17 pKa = 9.78EE18 pKa = 4.68FPDD21 pKa = 4.55KK22 pKa = 10.98INTEE26 pKa = 3.61IRR28 pKa = 11.84EE29 pKa = 4.0FSHH32 pKa = 7.14NIVDD36 pKa = 4.52KK37 pKa = 10.88IRR39 pKa = 11.84YY40 pKa = 6.44IWVWLVVIVLLLLIIKK56 pKa = 7.72ITPLLVNLVISCKK69 pKa = 9.83RR70 pKa = 11.84CYY72 pKa = 10.47QNFITGSKK80 pKa = 9.45KK81 pKa = 10.42LKK83 pKa = 10.35EE84 pKa = 4.43EE85 pKa = 3.99EE86 pKa = 4.21TEE88 pKa = 4.06IAA90 pKa = 4.37
Molecular weight: 10.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4810 |
90 |
2128 |
437.3 |
50.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.807 ± 0.349 | 1.996 ± 0.182 |
5.904 ± 0.348 | 7.173 ± 0.362 |
3.867 ± 0.203 | 6.071 ± 0.312 |
1.809 ± 0.188 | 8.919 ± 0.699 |
8.898 ± 0.402 | 9.376 ± 0.746 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.035 ± 0.368 | 7.006 ± 0.407 |
3.389 ± 0.226 | 2.495 ± 0.152 |
4.262 ± 0.323 | 6.736 ± 0.499 |
4.99 ± 0.308 | 5.301 ± 0.257 |
1.746 ± 0.17 | 4.22 ± 0.44 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
