
Mangrovimonas sp. DI 80
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Mangrovimonas; unclassified Mangrovimonas
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
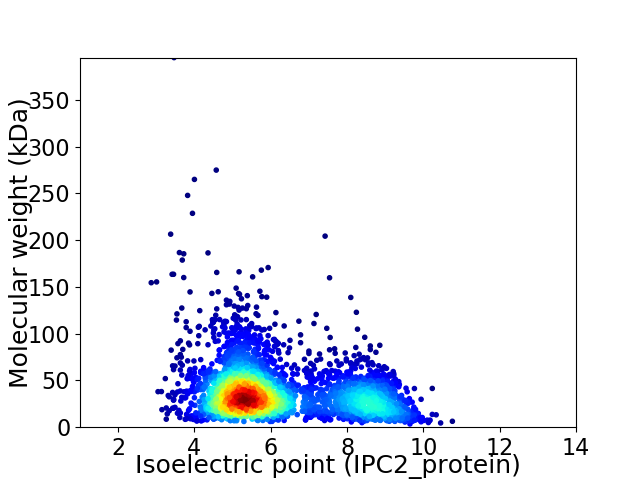
Virtual 2D-PAGE plot for 3194 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1V1ZJV8|A0A1V1ZJV8_9FLAO Sialidase domain-containing protein OS=Mangrovimonas sp. DI 80 OX=1779330 GN=BKM32_04820 PE=4 SV=1
MM1 pKa = 7.84RR2 pKa = 11.84KK3 pKa = 9.65FSFQLLVLSLVLFAVGCSNDD23 pKa = 3.26SSDD26 pKa = 3.71VEE28 pKa = 4.57EE29 pKa = 4.48IQVTSITILGEE40 pKa = 4.22TISDD44 pKa = 3.76GLSSQLSVQIVPANATEE61 pKa = 4.36TSVTWSVSDD70 pKa = 3.54PTVATISNNGLLSAVSNGSVNVTATSQDD98 pKa = 3.03GSGVFGDD105 pKa = 3.86RR106 pKa = 11.84SFTISGVGVGPVIVVQTSQEE126 pKa = 3.67ITEE129 pKa = 4.81AIITASAGDD138 pKa = 3.93IIYY141 pKa = 10.8VKK143 pKa = 10.57GGEE146 pKa = 4.06YY147 pKa = 10.48QFASTINFSADD158 pKa = 3.08GTSEE162 pKa = 3.74NAIIFMGDD170 pKa = 3.21PDD172 pKa = 4.52DD173 pKa = 4.91SEE175 pKa = 5.37RR176 pKa = 11.84PQFNFSAMSEE186 pKa = 4.04NSSNRR191 pKa = 11.84GIQLSGDD198 pKa = 3.16YY199 pKa = 9.18WHH201 pKa = 7.49IKK203 pKa = 9.74GIDD206 pKa = 3.36VFGAGDD212 pKa = 3.45NGMYY216 pKa = 8.7ITGHH220 pKa = 5.51YY221 pKa = 10.21NLVEE225 pKa = 4.06FCTFSEE231 pKa = 4.38NADD234 pKa = 3.41SGLQISGGGSNNTILNCDD252 pKa = 3.07SFYY255 pKa = 11.34NADD258 pKa = 3.7SSLEE262 pKa = 4.04NADD265 pKa = 4.03GFACKK270 pKa = 9.11LTAGTDD276 pKa = 3.34NKK278 pKa = 10.68FIGCRR283 pKa = 11.84AWQNLDD289 pKa = 4.34DD290 pKa = 4.74GWDD293 pKa = 3.59GYY295 pKa = 11.67LRR297 pKa = 11.84DD298 pKa = 4.16NDD300 pKa = 4.62NITTYY305 pKa = 11.25YY306 pKa = 8.36EE307 pKa = 3.95NCWAFKK313 pKa = 10.51NGYY316 pKa = 10.17LMDD319 pKa = 4.64GSAGIGDD326 pKa = 3.84GNGFKK331 pKa = 10.23TGGSDD336 pKa = 4.36DD337 pKa = 3.81KK338 pKa = 10.88TLKK341 pKa = 10.1HH342 pKa = 6.1HH343 pKa = 6.78GVYY346 pKa = 10.28KK347 pKa = 10.71NCIAAGNIYY356 pKa = 10.62DD357 pKa = 4.78GFDD360 pKa = 3.38HH361 pKa = 6.81NSNRR365 pKa = 11.84GDD367 pKa = 3.34IEE369 pKa = 4.89LYY371 pKa = 10.58NCSAYY376 pKa = 11.18NNGRR380 pKa = 11.84NISFSSTNIANFLLIKK396 pKa = 10.61NSLSFEE402 pKa = 4.46GNNSDD407 pKa = 5.01SYY409 pKa = 11.73SATQTDD415 pKa = 3.15ISNNGWQSGLTTDD428 pKa = 4.2ASDD431 pKa = 4.25FVSLDD436 pKa = 3.22MDD438 pKa = 4.66LLASPRR444 pKa = 11.84NADD447 pKa = 3.32GSLPDD452 pKa = 3.77IDD454 pKa = 4.65FLKK457 pKa = 10.76LSSGSDD463 pKa = 3.78LIDD466 pKa = 3.25QGVDD470 pKa = 2.82VGLPFLGTAPDD481 pKa = 3.3IGAFEE486 pKa = 4.18YY487 pKa = 10.9QDD489 pKa = 3.15
MM1 pKa = 7.84RR2 pKa = 11.84KK3 pKa = 9.65FSFQLLVLSLVLFAVGCSNDD23 pKa = 3.26SSDD26 pKa = 3.71VEE28 pKa = 4.57EE29 pKa = 4.48IQVTSITILGEE40 pKa = 4.22TISDD44 pKa = 3.76GLSSQLSVQIVPANATEE61 pKa = 4.36TSVTWSVSDD70 pKa = 3.54PTVATISNNGLLSAVSNGSVNVTATSQDD98 pKa = 3.03GSGVFGDD105 pKa = 3.86RR106 pKa = 11.84SFTISGVGVGPVIVVQTSQEE126 pKa = 3.67ITEE129 pKa = 4.81AIITASAGDD138 pKa = 3.93IIYY141 pKa = 10.8VKK143 pKa = 10.57GGEE146 pKa = 4.06YY147 pKa = 10.48QFASTINFSADD158 pKa = 3.08GTSEE162 pKa = 3.74NAIIFMGDD170 pKa = 3.21PDD172 pKa = 4.52DD173 pKa = 4.91SEE175 pKa = 5.37RR176 pKa = 11.84PQFNFSAMSEE186 pKa = 4.04NSSNRR191 pKa = 11.84GIQLSGDD198 pKa = 3.16YY199 pKa = 9.18WHH201 pKa = 7.49IKK203 pKa = 9.74GIDD206 pKa = 3.36VFGAGDD212 pKa = 3.45NGMYY216 pKa = 8.7ITGHH220 pKa = 5.51YY221 pKa = 10.21NLVEE225 pKa = 4.06FCTFSEE231 pKa = 4.38NADD234 pKa = 3.41SGLQISGGGSNNTILNCDD252 pKa = 3.07SFYY255 pKa = 11.34NADD258 pKa = 3.7SSLEE262 pKa = 4.04NADD265 pKa = 4.03GFACKK270 pKa = 9.11LTAGTDD276 pKa = 3.34NKK278 pKa = 10.68FIGCRR283 pKa = 11.84AWQNLDD289 pKa = 4.34DD290 pKa = 4.74GWDD293 pKa = 3.59GYY295 pKa = 11.67LRR297 pKa = 11.84DD298 pKa = 4.16NDD300 pKa = 4.62NITTYY305 pKa = 11.25YY306 pKa = 8.36EE307 pKa = 3.95NCWAFKK313 pKa = 10.51NGYY316 pKa = 10.17LMDD319 pKa = 4.64GSAGIGDD326 pKa = 3.84GNGFKK331 pKa = 10.23TGGSDD336 pKa = 4.36DD337 pKa = 3.81KK338 pKa = 10.88TLKK341 pKa = 10.1HH342 pKa = 6.1HH343 pKa = 6.78GVYY346 pKa = 10.28KK347 pKa = 10.71NCIAAGNIYY356 pKa = 10.62DD357 pKa = 4.78GFDD360 pKa = 3.38HH361 pKa = 6.81NSNRR365 pKa = 11.84GDD367 pKa = 3.34IEE369 pKa = 4.89LYY371 pKa = 10.58NCSAYY376 pKa = 11.18NNGRR380 pKa = 11.84NISFSSTNIANFLLIKK396 pKa = 10.61NSLSFEE402 pKa = 4.46GNNSDD407 pKa = 5.01SYY409 pKa = 11.73SATQTDD415 pKa = 3.15ISNNGWQSGLTTDD428 pKa = 4.2ASDD431 pKa = 4.25FVSLDD436 pKa = 3.22MDD438 pKa = 4.66LLASPRR444 pKa = 11.84NADD447 pKa = 3.32GSLPDD452 pKa = 3.77IDD454 pKa = 4.65FLKK457 pKa = 10.76LSSGSDD463 pKa = 3.78LIDD466 pKa = 3.25QGVDD470 pKa = 2.82VGLPFLGTAPDD481 pKa = 3.3IGAFEE486 pKa = 4.18YY487 pKa = 10.9QDD489 pKa = 3.15
Molecular weight: 52.0 kDa
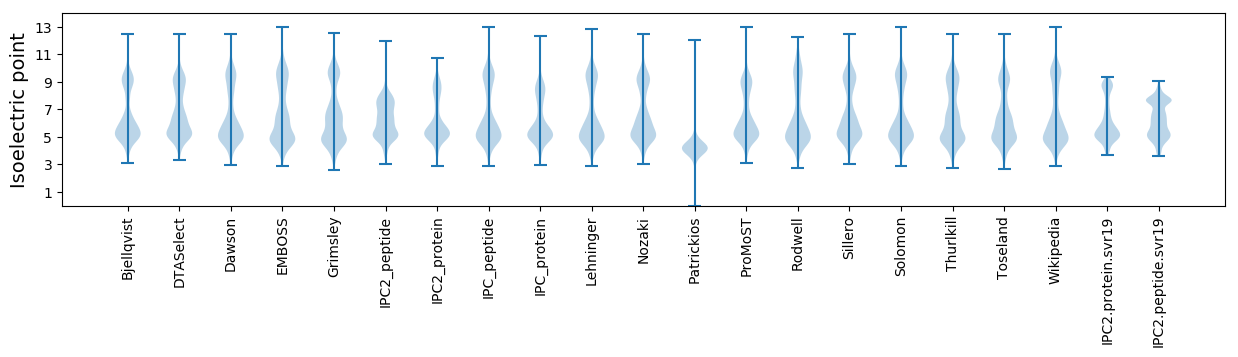
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V1ZGM4|A0A1V1ZGM4_9FLAO RNA-binding protein OS=Mangrovimonas sp. DI 80 OX=1779330 GN=BKM32_09690 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.52LSVSSEE48 pKa = 3.87PRR50 pKa = 11.84HH51 pKa = 5.92KK52 pKa = 10.61KK53 pKa = 9.84
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.52LSVSSEE48 pKa = 3.87PRR50 pKa = 11.84HH51 pKa = 5.92KK52 pKa = 10.61KK53 pKa = 9.84
Molecular weight: 6.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1127307 |
30 |
3754 |
352.9 |
39.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.531 ± 0.044 | 0.789 ± 0.015 |
5.679 ± 0.029 | 6.762 ± 0.038 |
5.156 ± 0.033 | 6.531 ± 0.04 |
1.891 ± 0.02 | 7.418 ± 0.04 |
7.109 ± 0.057 | 9.384 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.299 ± 0.023 | 5.916 ± 0.045 |
3.554 ± 0.026 | 3.504 ± 0.024 |
3.297 ± 0.03 | 6.507 ± 0.036 |
5.915 ± 0.06 | 6.436 ± 0.035 |
1.091 ± 0.016 | 4.232 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
