
Hubei sobemo-like virus 28
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
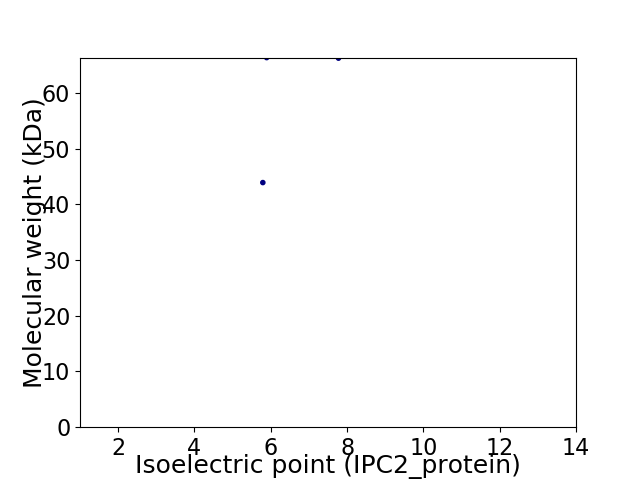
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
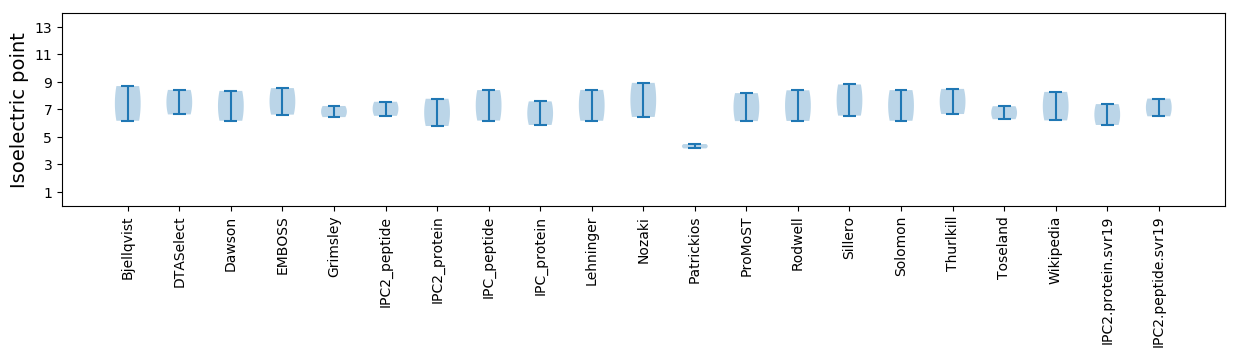
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEJ3|A0A1L3KEJ3_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 28 OX=1923214 PE=4 SV=1
MM1 pKa = 7.58GEE3 pKa = 4.35AIIDD7 pKa = 3.59RR8 pKa = 11.84AVQQHH13 pKa = 6.25RR14 pKa = 11.84SARR17 pKa = 11.84WVIPEE22 pKa = 4.11DD23 pKa = 4.39FISRR27 pKa = 11.84AHH29 pKa = 7.11FDD31 pKa = 3.29RR32 pKa = 11.84VLQKK36 pKa = 10.99LDD38 pKa = 3.5MQSSPGYY45 pKa = 8.33PYY47 pKa = 10.58CVRR50 pKa = 11.84IPVNAHH56 pKa = 6.14LFGVNADD63 pKa = 3.95GEE65 pKa = 4.35KK66 pKa = 11.05DD67 pKa = 3.18PDD69 pKa = 3.41AVEE72 pKa = 4.4RR73 pKa = 11.84VWEE76 pKa = 4.21AVQHH80 pKa = 6.04RR81 pKa = 11.84LGKK84 pKa = 10.55LLDD87 pKa = 4.04GEE89 pKa = 4.55EE90 pKa = 4.37DD91 pKa = 3.75CDD93 pKa = 5.25PIRR96 pKa = 11.84LFIKK100 pKa = 10.27PEE102 pKa = 3.73PLKK105 pKa = 10.56EE106 pKa = 4.15KK107 pKa = 10.61KK108 pKa = 9.26LTEE111 pKa = 3.48HH112 pKa = 7.77RR113 pKa = 11.84YY114 pKa = 10.01RR115 pKa = 11.84IISSVSIVDD124 pKa = 3.61QIIDD128 pKa = 3.66HH129 pKa = 7.01LLFDD133 pKa = 5.29DD134 pKa = 4.59MNEE137 pKa = 3.65TMYY140 pKa = 11.44ANWHH144 pKa = 5.8NVPSMVGWSPFVGGWKK160 pKa = 10.17VMPKK164 pKa = 10.32LKK166 pKa = 9.92WLAIDD171 pKa = 3.63KK172 pKa = 10.74SSWDD176 pKa = 3.36WTVHH180 pKa = 5.71KK181 pKa = 10.13WILDD185 pKa = 3.32QCLEE189 pKa = 4.07LRR191 pKa = 11.84TRR193 pKa = 11.84LCDD196 pKa = 3.55NMNAAWLKK204 pKa = 9.66LARR207 pKa = 11.84ARR209 pKa = 11.84YY210 pKa = 8.46HH211 pKa = 6.82LLYY214 pKa = 10.63DD215 pKa = 3.58NPVLVTSGGVLLRR228 pKa = 11.84QKK230 pKa = 9.53TPGVMKK236 pKa = 10.13SGCVNTIADD245 pKa = 3.82NSIAQYY251 pKa = 11.38LLDD254 pKa = 4.13LRR256 pKa = 11.84VCFEE260 pKa = 4.14IGLTPGYY267 pKa = 9.86LMSMGDD273 pKa = 3.76DD274 pKa = 3.56TLQQKK279 pKa = 9.57PRR281 pKa = 11.84DD282 pKa = 3.66LQKK285 pKa = 11.29YY286 pKa = 9.68LDD288 pKa = 4.48ALSSFCIVKK297 pKa = 10.09QATEE301 pKa = 3.97ANEE304 pKa = 3.82FAGMLFNGRR313 pKa = 11.84NVEE316 pKa = 3.89PLYY319 pKa = 10.58HH320 pKa = 6.71GKK322 pKa = 8.97HH323 pKa = 5.84ACTILHH329 pKa = 7.07AKK331 pKa = 10.15PEE333 pKa = 4.17ILEE336 pKa = 4.2EE337 pKa = 4.02LADD340 pKa = 4.62AYY342 pKa = 9.88TLLYY346 pKa = 10.22HH347 pKa = 7.14RR348 pKa = 11.84SKK350 pKa = 10.48FRR352 pKa = 11.84NWFEE356 pKa = 4.12NLFTAMGIDD365 pKa = 3.79FHH367 pKa = 6.69PRR369 pKa = 11.84WKK371 pKa = 10.23RR372 pKa = 11.84DD373 pKa = 3.53AIYY376 pKa = 10.74DD377 pKa = 3.93GEE379 pKa = 4.33DD380 pKa = 2.86
MM1 pKa = 7.58GEE3 pKa = 4.35AIIDD7 pKa = 3.59RR8 pKa = 11.84AVQQHH13 pKa = 6.25RR14 pKa = 11.84SARR17 pKa = 11.84WVIPEE22 pKa = 4.11DD23 pKa = 4.39FISRR27 pKa = 11.84AHH29 pKa = 7.11FDD31 pKa = 3.29RR32 pKa = 11.84VLQKK36 pKa = 10.99LDD38 pKa = 3.5MQSSPGYY45 pKa = 8.33PYY47 pKa = 10.58CVRR50 pKa = 11.84IPVNAHH56 pKa = 6.14LFGVNADD63 pKa = 3.95GEE65 pKa = 4.35KK66 pKa = 11.05DD67 pKa = 3.18PDD69 pKa = 3.41AVEE72 pKa = 4.4RR73 pKa = 11.84VWEE76 pKa = 4.21AVQHH80 pKa = 6.04RR81 pKa = 11.84LGKK84 pKa = 10.55LLDD87 pKa = 4.04GEE89 pKa = 4.55EE90 pKa = 4.37DD91 pKa = 3.75CDD93 pKa = 5.25PIRR96 pKa = 11.84LFIKK100 pKa = 10.27PEE102 pKa = 3.73PLKK105 pKa = 10.56EE106 pKa = 4.15KK107 pKa = 10.61KK108 pKa = 9.26LTEE111 pKa = 3.48HH112 pKa = 7.77RR113 pKa = 11.84YY114 pKa = 10.01RR115 pKa = 11.84IISSVSIVDD124 pKa = 3.61QIIDD128 pKa = 3.66HH129 pKa = 7.01LLFDD133 pKa = 5.29DD134 pKa = 4.59MNEE137 pKa = 3.65TMYY140 pKa = 11.44ANWHH144 pKa = 5.8NVPSMVGWSPFVGGWKK160 pKa = 10.17VMPKK164 pKa = 10.32LKK166 pKa = 9.92WLAIDD171 pKa = 3.63KK172 pKa = 10.74SSWDD176 pKa = 3.36WTVHH180 pKa = 5.71KK181 pKa = 10.13WILDD185 pKa = 3.32QCLEE189 pKa = 4.07LRR191 pKa = 11.84TRR193 pKa = 11.84LCDD196 pKa = 3.55NMNAAWLKK204 pKa = 9.66LARR207 pKa = 11.84ARR209 pKa = 11.84YY210 pKa = 8.46HH211 pKa = 6.82LLYY214 pKa = 10.63DD215 pKa = 3.58NPVLVTSGGVLLRR228 pKa = 11.84QKK230 pKa = 9.53TPGVMKK236 pKa = 10.13SGCVNTIADD245 pKa = 3.82NSIAQYY251 pKa = 11.38LLDD254 pKa = 4.13LRR256 pKa = 11.84VCFEE260 pKa = 4.14IGLTPGYY267 pKa = 9.86LMSMGDD273 pKa = 3.76DD274 pKa = 3.56TLQQKK279 pKa = 9.57PRR281 pKa = 11.84DD282 pKa = 3.66LQKK285 pKa = 11.29YY286 pKa = 9.68LDD288 pKa = 4.48ALSSFCIVKK297 pKa = 10.09QATEE301 pKa = 3.97ANEE304 pKa = 3.82FAGMLFNGRR313 pKa = 11.84NVEE316 pKa = 3.89PLYY319 pKa = 10.58HH320 pKa = 6.71GKK322 pKa = 8.97HH323 pKa = 5.84ACTILHH329 pKa = 7.07AKK331 pKa = 10.15PEE333 pKa = 4.17ILEE336 pKa = 4.2EE337 pKa = 4.02LADD340 pKa = 4.62AYY342 pKa = 9.88TLLYY346 pKa = 10.22HH347 pKa = 7.14RR348 pKa = 11.84SKK350 pKa = 10.48FRR352 pKa = 11.84NWFEE356 pKa = 4.12NLFTAMGIDD365 pKa = 3.79FHH367 pKa = 6.69PRR369 pKa = 11.84WKK371 pKa = 10.23RR372 pKa = 11.84DD373 pKa = 3.53AIYY376 pKa = 10.74DD377 pKa = 3.93GEE379 pKa = 4.33DD380 pKa = 2.86
Molecular weight: 43.89 kDa
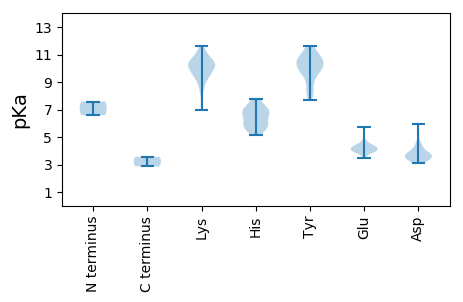
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEJ3|A0A1L3KEJ3_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 28 OX=1923214 PE=4 SV=1
MM1 pKa = 6.58TTNSEE6 pKa = 4.1QIYY9 pKa = 10.16NPLSEE14 pKa = 4.53EE15 pKa = 4.06VEE17 pKa = 4.04AQPRR21 pKa = 11.84CGSWLSRR28 pKa = 11.84ALKK31 pKa = 9.24WVLAVLVAAVTVPLIWSAFCGLMIGLSGWVLGWYY65 pKa = 7.71RR66 pKa = 11.84TQHH69 pKa = 5.5LALYY73 pKa = 8.82EE74 pKa = 3.97LAKK77 pKa = 10.73NAFGPPPSPPEE88 pKa = 3.93PDD90 pKa = 3.4VVEE93 pKa = 4.85QYY95 pKa = 10.66AQKK98 pKa = 11.03LLDD101 pKa = 3.52FAEE104 pKa = 4.72RR105 pKa = 11.84VDD107 pKa = 3.87STLLATLLVAIIMVISIWITFGLLMRR133 pKa = 11.84SLRR136 pKa = 11.84RR137 pKa = 11.84ISYY140 pKa = 9.35KK141 pKa = 10.42LRR143 pKa = 11.84GIHH146 pKa = 6.51LEE148 pKa = 3.94SMQPGSDD155 pKa = 3.5FTASDD160 pKa = 3.39IPSYY164 pKa = 8.08QVKK167 pKa = 10.08VLSPGLLGDD176 pKa = 3.56SHH178 pKa = 7.76QGYY181 pKa = 9.68GIRR184 pKa = 11.84VGSWLVVPLHH194 pKa = 5.82VLNSVSEE201 pKa = 4.23LMLQGPTGQKK211 pKa = 9.03MVMRR215 pKa = 11.84KK216 pKa = 8.3QYY218 pKa = 10.21EE219 pKa = 4.23VSRR222 pKa = 11.84LHH224 pKa = 7.25ADD226 pKa = 3.13VAYY229 pKa = 10.55LYY231 pKa = 10.72LEE233 pKa = 4.29KK234 pKa = 10.68EE235 pKa = 4.13QWSKK239 pKa = 11.56LGAKK243 pKa = 8.55NAKK246 pKa = 6.94TASMCRR252 pKa = 11.84GFARR256 pKa = 11.84CYY258 pKa = 10.19GPRR261 pKa = 11.84GYY263 pKa = 11.65SMGSMDD269 pKa = 4.12RR270 pKa = 11.84SGLFGMLVYY279 pKa = 10.57SGSTIPGMSGAAYY292 pKa = 9.6EE293 pKa = 4.34LMGKK297 pKa = 9.41VVGLHH302 pKa = 5.78TGCTNTTNMGVSAALFVEE320 pKa = 4.36EE321 pKa = 5.76LMTKK325 pKa = 9.43TPLKK329 pKa = 10.27EE330 pKa = 3.99EE331 pKa = 4.17SASALGISSGVGEE344 pKa = 4.23QSAAVYY350 pKa = 10.8SLGSKK355 pKa = 9.58IGKK358 pKa = 9.37QIEE361 pKa = 4.25AWDD364 pKa = 3.5KK365 pKa = 10.36AQIKK369 pKa = 9.61EE370 pKa = 4.12RR371 pKa = 11.84LANPDD376 pKa = 3.08SRR378 pKa = 11.84PWHH381 pKa = 6.65EE382 pKa = 5.42DD383 pKa = 3.11DD384 pKa = 5.97EE385 pKa = 5.67IDD387 pKa = 3.88YY388 pKa = 11.0DD389 pKa = 3.7QDD391 pKa = 3.9LGLEE395 pKa = 4.37SASKK399 pKa = 10.07PKK401 pKa = 10.52KK402 pKa = 10.02LVPLGIVQQPLLLQSDD418 pKa = 4.24SPKK421 pKa = 11.11EE422 pKa = 4.0EE423 pKa = 4.12VLTLADD429 pKa = 3.7VTGLAARR436 pKa = 11.84VTALEE441 pKa = 3.95EE442 pKa = 4.38CVRR445 pKa = 11.84EE446 pKa = 4.03LQQLAIKK453 pKa = 10.18KK454 pKa = 9.55KK455 pKa = 10.36AEE457 pKa = 3.94KK458 pKa = 10.5QEE460 pKa = 4.59FKK462 pKa = 11.29CPDD465 pKa = 3.51CDD467 pKa = 3.35IACRR471 pKa = 11.84TEE473 pKa = 4.35LRR475 pKa = 11.84LSNHH479 pKa = 6.66LKK481 pKa = 8.28FTCAKK486 pKa = 9.54RR487 pKa = 11.84VSFDD491 pKa = 4.43CEE493 pKa = 3.93LCEE496 pKa = 4.46TKK498 pKa = 10.59CRR500 pKa = 11.84TAEE503 pKa = 3.81TLKK506 pKa = 10.06RR507 pKa = 11.84HH508 pKa = 5.4MVNSHH513 pKa = 5.17TKK515 pKa = 9.54VKK517 pKa = 10.5IEE519 pKa = 4.35GEE521 pKa = 4.0SAYY524 pKa = 10.01PGDD527 pKa = 3.9SKK529 pKa = 11.6VVVQTGPFLAKK540 pKa = 10.04RR541 pKa = 11.84RR542 pKa = 11.84ASQKK546 pKa = 9.73GRR548 pKa = 11.84KK549 pKa = 7.32KK550 pKa = 8.65TSSRR554 pKa = 11.84SSSSSGDD561 pKa = 3.22RR562 pKa = 11.84SRR564 pKa = 11.84SPSLEE569 pKa = 3.87EE570 pKa = 3.87TLSLMTSSLLRR581 pKa = 11.84IEE583 pKa = 4.69KK584 pKa = 10.34SLNKK588 pKa = 10.13NQGDD592 pKa = 3.8TAGQSLATMLNN603 pKa = 3.57
MM1 pKa = 6.58TTNSEE6 pKa = 4.1QIYY9 pKa = 10.16NPLSEE14 pKa = 4.53EE15 pKa = 4.06VEE17 pKa = 4.04AQPRR21 pKa = 11.84CGSWLSRR28 pKa = 11.84ALKK31 pKa = 9.24WVLAVLVAAVTVPLIWSAFCGLMIGLSGWVLGWYY65 pKa = 7.71RR66 pKa = 11.84TQHH69 pKa = 5.5LALYY73 pKa = 8.82EE74 pKa = 3.97LAKK77 pKa = 10.73NAFGPPPSPPEE88 pKa = 3.93PDD90 pKa = 3.4VVEE93 pKa = 4.85QYY95 pKa = 10.66AQKK98 pKa = 11.03LLDD101 pKa = 3.52FAEE104 pKa = 4.72RR105 pKa = 11.84VDD107 pKa = 3.87STLLATLLVAIIMVISIWITFGLLMRR133 pKa = 11.84SLRR136 pKa = 11.84RR137 pKa = 11.84ISYY140 pKa = 9.35KK141 pKa = 10.42LRR143 pKa = 11.84GIHH146 pKa = 6.51LEE148 pKa = 3.94SMQPGSDD155 pKa = 3.5FTASDD160 pKa = 3.39IPSYY164 pKa = 8.08QVKK167 pKa = 10.08VLSPGLLGDD176 pKa = 3.56SHH178 pKa = 7.76QGYY181 pKa = 9.68GIRR184 pKa = 11.84VGSWLVVPLHH194 pKa = 5.82VLNSVSEE201 pKa = 4.23LMLQGPTGQKK211 pKa = 9.03MVMRR215 pKa = 11.84KK216 pKa = 8.3QYY218 pKa = 10.21EE219 pKa = 4.23VSRR222 pKa = 11.84LHH224 pKa = 7.25ADD226 pKa = 3.13VAYY229 pKa = 10.55LYY231 pKa = 10.72LEE233 pKa = 4.29KK234 pKa = 10.68EE235 pKa = 4.13QWSKK239 pKa = 11.56LGAKK243 pKa = 8.55NAKK246 pKa = 6.94TASMCRR252 pKa = 11.84GFARR256 pKa = 11.84CYY258 pKa = 10.19GPRR261 pKa = 11.84GYY263 pKa = 11.65SMGSMDD269 pKa = 4.12RR270 pKa = 11.84SGLFGMLVYY279 pKa = 10.57SGSTIPGMSGAAYY292 pKa = 9.6EE293 pKa = 4.34LMGKK297 pKa = 9.41VVGLHH302 pKa = 5.78TGCTNTTNMGVSAALFVEE320 pKa = 4.36EE321 pKa = 5.76LMTKK325 pKa = 9.43TPLKK329 pKa = 10.27EE330 pKa = 3.99EE331 pKa = 4.17SASALGISSGVGEE344 pKa = 4.23QSAAVYY350 pKa = 10.8SLGSKK355 pKa = 9.58IGKK358 pKa = 9.37QIEE361 pKa = 4.25AWDD364 pKa = 3.5KK365 pKa = 10.36AQIKK369 pKa = 9.61EE370 pKa = 4.12RR371 pKa = 11.84LANPDD376 pKa = 3.08SRR378 pKa = 11.84PWHH381 pKa = 6.65EE382 pKa = 5.42DD383 pKa = 3.11DD384 pKa = 5.97EE385 pKa = 5.67IDD387 pKa = 3.88YY388 pKa = 11.0DD389 pKa = 3.7QDD391 pKa = 3.9LGLEE395 pKa = 4.37SASKK399 pKa = 10.07PKK401 pKa = 10.52KK402 pKa = 10.02LVPLGIVQQPLLLQSDD418 pKa = 4.24SPKK421 pKa = 11.11EE422 pKa = 4.0EE423 pKa = 4.12VLTLADD429 pKa = 3.7VTGLAARR436 pKa = 11.84VTALEE441 pKa = 3.95EE442 pKa = 4.38CVRR445 pKa = 11.84EE446 pKa = 4.03LQQLAIKK453 pKa = 10.18KK454 pKa = 9.55KK455 pKa = 10.36AEE457 pKa = 3.94KK458 pKa = 10.5QEE460 pKa = 4.59FKK462 pKa = 11.29CPDD465 pKa = 3.51CDD467 pKa = 3.35IACRR471 pKa = 11.84TEE473 pKa = 4.35LRR475 pKa = 11.84LSNHH479 pKa = 6.66LKK481 pKa = 8.28FTCAKK486 pKa = 9.54RR487 pKa = 11.84VSFDD491 pKa = 4.43CEE493 pKa = 3.93LCEE496 pKa = 4.46TKK498 pKa = 10.59CRR500 pKa = 11.84TAEE503 pKa = 3.81TLKK506 pKa = 10.06RR507 pKa = 11.84HH508 pKa = 5.4MVNSHH513 pKa = 5.17TKK515 pKa = 9.54VKK517 pKa = 10.5IEE519 pKa = 4.35GEE521 pKa = 4.0SAYY524 pKa = 10.01PGDD527 pKa = 3.9SKK529 pKa = 11.6VVVQTGPFLAKK540 pKa = 10.04RR541 pKa = 11.84RR542 pKa = 11.84ASQKK546 pKa = 9.73GRR548 pKa = 11.84KK549 pKa = 7.32KK550 pKa = 8.65TSSRR554 pKa = 11.84SSSSSGDD561 pKa = 3.22RR562 pKa = 11.84SRR564 pKa = 11.84SPSLEE569 pKa = 3.87EE570 pKa = 3.87TLSLMTSSLLRR581 pKa = 11.84IEE583 pKa = 4.69KK584 pKa = 10.34SLNKK588 pKa = 10.13NQGDD592 pKa = 3.8TAGQSLATMLNN603 pKa = 3.57
Molecular weight: 66.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
983 |
380 |
603 |
491.5 |
55.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.325 ± 0.279 | 2.136 ± 0.018 |
5.392 ± 1.45 | 6.205 ± 0.393 |
2.645 ± 0.602 | 6.714 ± 0.688 |
2.442 ± 0.72 | 4.578 ± 0.702 |
6.307 ± 0.3 | 11.495 ± 0.409 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.154 ± 0.002 | 2.848 ± 0.637 |
4.578 ± 0.092 | 3.967 ± 0.317 |
5.493 ± 0.324 | 8.24 ± 2.029 |
4.578 ± 0.67 | 6.612 ± 0.172 |
2.238 ± 0.533 | 3.052 ± 0.214 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
