
Plasmodium berghei (strain Anka)
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Apicomplexa; Aconoidasida; Haemosporida; Plasmodiidae; Plasmodium; Plasmodium (Vinckeia); Plasmodium berghei
Average proteome isoelectric point is 7.38
Get precalculated fractions of proteins
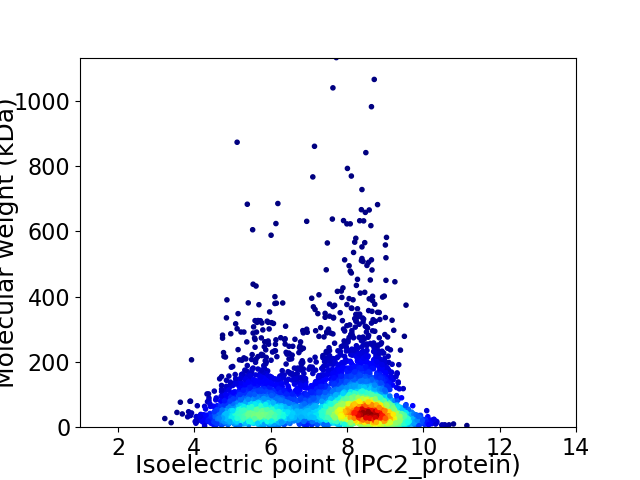
Virtual 2D-PAGE plot for 4928 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A509AEL7|A0A509AEL7_PLABA Cleavage and polyadenylation specificity factor subunit 5 OS=Plasmodium berghei (strain Anka) OX=5823 GN=PBANKA_0204400 PE=3 SV=1
MM1 pKa = 7.68KK2 pKa = 10.54NNIMKK7 pKa = 10.15ISFALLFLSSYY18 pKa = 10.39FSEE21 pKa = 4.22INGNVTNKK29 pKa = 9.93NKK31 pKa = 9.88YY32 pKa = 8.92RR33 pKa = 11.84LRR35 pKa = 11.84SYY37 pKa = 8.98LTKK40 pKa = 10.85NEE42 pKa = 4.16DD43 pKa = 4.5LNNTSYY49 pKa = 11.27NGNIHH54 pKa = 6.63GEE56 pKa = 4.21SEE58 pKa = 4.2DD59 pKa = 4.2AISVGDD65 pKa = 3.62TTQTLKK71 pKa = 11.03NGEE74 pKa = 4.51SGTQGSSNLPQGKK87 pKa = 8.98SLQSTDD93 pKa = 3.59TNHH96 pKa = 6.11SQDD99 pKa = 2.57GHH101 pKa = 6.33TEE103 pKa = 4.01DD104 pKa = 4.56SCAEE108 pKa = 4.14CGRR111 pKa = 11.84GCTEE115 pKa = 3.92GCEE118 pKa = 4.48GCHH121 pKa = 6.66DD122 pKa = 4.57ASCLEE127 pKa = 4.1CHH129 pKa = 6.64EE130 pKa = 4.78GCEE133 pKa = 4.53DD134 pKa = 5.23CPDD137 pKa = 3.84CHH139 pKa = 7.71DD140 pKa = 4.79EE141 pKa = 4.81SCLDD145 pKa = 3.87CQTEE149 pKa = 4.26CTDD152 pKa = 3.24ACRR155 pKa = 11.84EE156 pKa = 3.85EE157 pKa = 4.29CAKK160 pKa = 10.55IKK162 pKa = 10.06TDD164 pKa = 3.25TVHH167 pKa = 7.51RR168 pKa = 11.84DD169 pKa = 3.28GEE171 pKa = 4.44NVQACTQCTTNDD183 pKa = 4.0PNCEE187 pKa = 3.97EE188 pKa = 4.25CKK190 pKa = 10.68KK191 pKa = 10.6IITTRR196 pKa = 11.84QPEE199 pKa = 4.43TPNSGIEE206 pKa = 3.98AEE208 pKa = 4.27PASVVQNAIEE218 pKa = 5.86DD219 pKa = 3.73ITNDD223 pKa = 3.58TKK225 pKa = 11.12TMPDD229 pKa = 3.39EE230 pKa = 4.44NEE232 pKa = 4.14AVCDD236 pKa = 3.78EE237 pKa = 4.13QLDD240 pKa = 3.81EE241 pKa = 5.56GGDD244 pKa = 3.67NEE246 pKa = 4.25EE247 pKa = 4.88EE248 pKa = 3.9EE249 pKa = 5.58DD250 pKa = 3.92IVDD253 pKa = 4.16SEE255 pKa = 4.68DD256 pKa = 3.48IEE258 pKa = 5.03IIEE261 pKa = 4.22HH262 pKa = 6.07TGDD265 pKa = 4.38IEE267 pKa = 5.59DD268 pKa = 4.41SDD270 pKa = 4.43DD271 pKa = 4.14SEE273 pKa = 4.34EE274 pKa = 5.09LEE276 pKa = 5.12LEE278 pKa = 4.31LEE280 pKa = 4.49LEE282 pKa = 4.2LEE284 pKa = 4.38EE285 pKa = 6.48AIDD288 pKa = 3.77MSRR291 pKa = 11.84DD292 pKa = 3.47GQEE295 pKa = 3.95EE296 pKa = 4.49NEE298 pKa = 4.59GEE300 pKa = 4.37TGTTTTNEE308 pKa = 3.48QRR310 pKa = 11.84NNHH313 pKa = 5.05NTNTNEE319 pKa = 3.94VTDD322 pKa = 3.97NNQTNNSNNTNNNNNGNHH340 pKa = 6.97FIIKK344 pKa = 9.34EE345 pKa = 3.96YY346 pKa = 10.57KK347 pKa = 9.8GNHH350 pKa = 5.6DD351 pKa = 4.05AKK353 pKa = 10.39TEE355 pKa = 3.97NEE357 pKa = 4.26NILNNALSDD366 pKa = 3.39IDD368 pKa = 5.29DD369 pKa = 4.02NQFSSDD375 pKa = 3.4IQDD378 pKa = 3.8FAQDD382 pKa = 3.0INEE385 pKa = 4.11YY386 pKa = 11.12VLMEE390 pKa = 4.23EE391 pKa = 5.59DD392 pKa = 3.1II393 pKa = 4.75
MM1 pKa = 7.68KK2 pKa = 10.54NNIMKK7 pKa = 10.15ISFALLFLSSYY18 pKa = 10.39FSEE21 pKa = 4.22INGNVTNKK29 pKa = 9.93NKK31 pKa = 9.88YY32 pKa = 8.92RR33 pKa = 11.84LRR35 pKa = 11.84SYY37 pKa = 8.98LTKK40 pKa = 10.85NEE42 pKa = 4.16DD43 pKa = 4.5LNNTSYY49 pKa = 11.27NGNIHH54 pKa = 6.63GEE56 pKa = 4.21SEE58 pKa = 4.2DD59 pKa = 4.2AISVGDD65 pKa = 3.62TTQTLKK71 pKa = 11.03NGEE74 pKa = 4.51SGTQGSSNLPQGKK87 pKa = 8.98SLQSTDD93 pKa = 3.59TNHH96 pKa = 6.11SQDD99 pKa = 2.57GHH101 pKa = 6.33TEE103 pKa = 4.01DD104 pKa = 4.56SCAEE108 pKa = 4.14CGRR111 pKa = 11.84GCTEE115 pKa = 3.92GCEE118 pKa = 4.48GCHH121 pKa = 6.66DD122 pKa = 4.57ASCLEE127 pKa = 4.1CHH129 pKa = 6.64EE130 pKa = 4.78GCEE133 pKa = 4.53DD134 pKa = 5.23CPDD137 pKa = 3.84CHH139 pKa = 7.71DD140 pKa = 4.79EE141 pKa = 4.81SCLDD145 pKa = 3.87CQTEE149 pKa = 4.26CTDD152 pKa = 3.24ACRR155 pKa = 11.84EE156 pKa = 3.85EE157 pKa = 4.29CAKK160 pKa = 10.55IKK162 pKa = 10.06TDD164 pKa = 3.25TVHH167 pKa = 7.51RR168 pKa = 11.84DD169 pKa = 3.28GEE171 pKa = 4.44NVQACTQCTTNDD183 pKa = 4.0PNCEE187 pKa = 3.97EE188 pKa = 4.25CKK190 pKa = 10.68KK191 pKa = 10.6IITTRR196 pKa = 11.84QPEE199 pKa = 4.43TPNSGIEE206 pKa = 3.98AEE208 pKa = 4.27PASVVQNAIEE218 pKa = 5.86DD219 pKa = 3.73ITNDD223 pKa = 3.58TKK225 pKa = 11.12TMPDD229 pKa = 3.39EE230 pKa = 4.44NEE232 pKa = 4.14AVCDD236 pKa = 3.78EE237 pKa = 4.13QLDD240 pKa = 3.81EE241 pKa = 5.56GGDD244 pKa = 3.67NEE246 pKa = 4.25EE247 pKa = 4.88EE248 pKa = 3.9EE249 pKa = 5.58DD250 pKa = 3.92IVDD253 pKa = 4.16SEE255 pKa = 4.68DD256 pKa = 3.48IEE258 pKa = 5.03IIEE261 pKa = 4.22HH262 pKa = 6.07TGDD265 pKa = 4.38IEE267 pKa = 5.59DD268 pKa = 4.41SDD270 pKa = 4.43DD271 pKa = 4.14SEE273 pKa = 4.34EE274 pKa = 5.09LEE276 pKa = 5.12LEE278 pKa = 4.31LEE280 pKa = 4.49LEE282 pKa = 4.2LEE284 pKa = 4.38EE285 pKa = 6.48AIDD288 pKa = 3.77MSRR291 pKa = 11.84DD292 pKa = 3.47GQEE295 pKa = 3.95EE296 pKa = 4.49NEE298 pKa = 4.59GEE300 pKa = 4.37TGTTTTNEE308 pKa = 3.48QRR310 pKa = 11.84NNHH313 pKa = 5.05NTNTNEE319 pKa = 3.94VTDD322 pKa = 3.97NNQTNNSNNTNNNNNGNHH340 pKa = 6.97FIIKK344 pKa = 9.34EE345 pKa = 3.96YY346 pKa = 10.57KK347 pKa = 9.8GNHH350 pKa = 5.6DD351 pKa = 4.05AKK353 pKa = 10.39TEE355 pKa = 3.97NEE357 pKa = 4.26NILNNALSDD366 pKa = 3.39IDD368 pKa = 5.29DD369 pKa = 4.02NQFSSDD375 pKa = 3.4IQDD378 pKa = 3.8FAQDD382 pKa = 3.0INEE385 pKa = 4.11YY386 pKa = 11.12VLMEE390 pKa = 4.23EE391 pKa = 5.59DD392 pKa = 3.1II393 pKa = 4.75
Molecular weight: 43.6 kDa
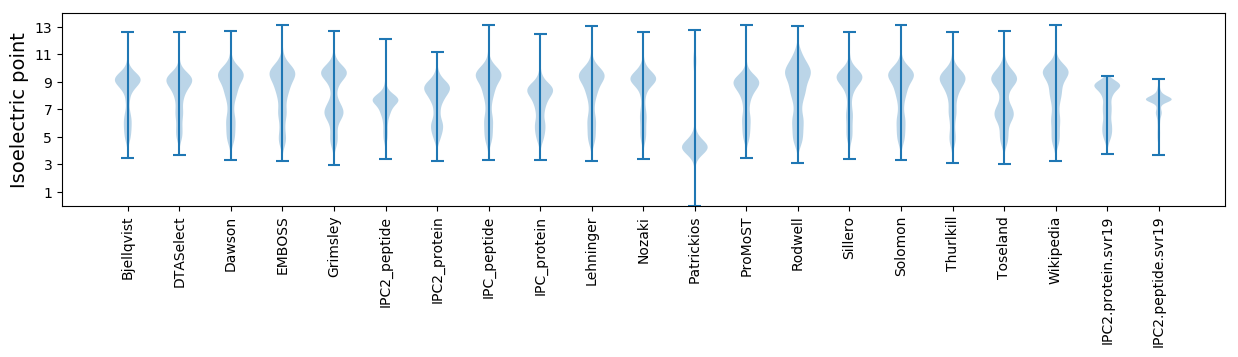
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A509AIL1|A0A509AIL1_PLABA AP2 domain transcription factor putative OS=Plasmodium berghei (strain Anka) OX=5823 GN=PBANKA_0932300 PE=4 SV=1
MM1 pKa = 7.28AHH3 pKa = 6.31GASRR7 pKa = 11.84YY8 pKa = 8.75KK9 pKa = 10.48KK10 pKa = 10.3SRR12 pKa = 11.84AKK14 pKa = 9.88MRR16 pKa = 11.84WKK18 pKa = 9.16WKK20 pKa = 9.29KK21 pKa = 9.72KK22 pKa = 6.67RR23 pKa = 11.84TRR25 pKa = 11.84RR26 pKa = 11.84LQKK29 pKa = 9.69KK30 pKa = 7.67RR31 pKa = 11.84RR32 pKa = 11.84KK33 pKa = 8.24MRR35 pKa = 11.84QRR37 pKa = 11.84SRR39 pKa = 3.22
MM1 pKa = 7.28AHH3 pKa = 6.31GASRR7 pKa = 11.84YY8 pKa = 8.75KK9 pKa = 10.48KK10 pKa = 10.3SRR12 pKa = 11.84AKK14 pKa = 9.88MRR16 pKa = 11.84WKK18 pKa = 9.16WKK20 pKa = 9.29KK21 pKa = 9.72KK22 pKa = 6.67RR23 pKa = 11.84TRR25 pKa = 11.84RR26 pKa = 11.84LQKK29 pKa = 9.69KK30 pKa = 7.67RR31 pKa = 11.84RR32 pKa = 11.84KK33 pKa = 8.24MRR35 pKa = 11.84QRR37 pKa = 11.84SRR39 pKa = 3.22
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3405970 |
29 |
9556 |
691.1 |
81.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.251 ± 0.018 | 1.702 ± 0.012 |
5.616 ± 0.023 | 7.093 ± 0.04 |
4.897 ± 0.03 | 3.007 ± 0.025 |
1.936 ± 0.011 | 10.203 ± 0.038 |
11.926 ± 0.039 | 7.976 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.884 ± 0.01 | 13.28 ± 0.07 |
2.086 ± 0.021 | 2.729 ± 0.017 |
2.557 ± 0.017 | 7.038 ± 0.03 |
4.203 ± 0.021 | 3.539 ± 0.018 |
0.497 ± 0.008 | 5.58 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
