
Human papillomavirus 126
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 11
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


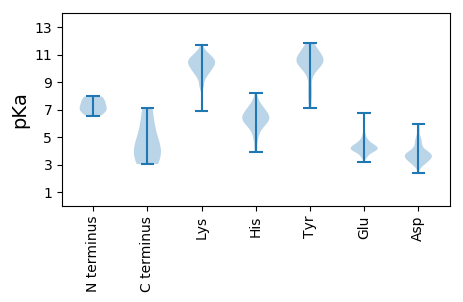
Protein with the lowest isoelectric point:
>tr|G5ELP1|G5ELP1_9PAPI Replication protein E1 OS=Human papillomavirus 126 OX=1055684 GN=HpV126gp3 PE=3 SV=1
MM1 pKa = 7.17MGKK4 pKa = 9.39EE5 pKa = 4.06CTLNDD10 pKa = 3.0IVLQEE15 pKa = 4.09NVEE18 pKa = 4.31DD19 pKa = 4.16LVLPAPLLCEE29 pKa = 4.05EE30 pKa = 4.84SSLSIDD36 pKa = 3.6DD37 pKa = 4.04TPEE40 pKa = 3.87EE41 pKa = 4.38EE42 pKa = 4.56LLSPYY47 pKa = 10.39RR48 pKa = 11.84VDD50 pKa = 4.92SYY52 pKa = 11.73CFYY55 pKa = 10.75CNRR58 pKa = 11.84GVRR61 pKa = 11.84LSVVASDD68 pKa = 3.61GAVYY72 pKa = 10.93LLQQLLLKK80 pKa = 10.41DD81 pKa = 3.64LTIVCPSCTRR91 pKa = 11.84QNLQNGRR98 pKa = 11.84SHH100 pKa = 7.1
MM1 pKa = 7.17MGKK4 pKa = 9.39EE5 pKa = 4.06CTLNDD10 pKa = 3.0IVLQEE15 pKa = 4.09NVEE18 pKa = 4.31DD19 pKa = 4.16LVLPAPLLCEE29 pKa = 4.05EE30 pKa = 4.84SSLSIDD36 pKa = 3.6DD37 pKa = 4.04TPEE40 pKa = 3.87EE41 pKa = 4.38EE42 pKa = 4.56LLSPYY47 pKa = 10.39RR48 pKa = 11.84VDD50 pKa = 4.92SYY52 pKa = 11.73CFYY55 pKa = 10.75CNRR58 pKa = 11.84GVRR61 pKa = 11.84LSVVASDD68 pKa = 3.61GAVYY72 pKa = 10.93LLQQLLLKK80 pKa = 10.41DD81 pKa = 3.64LTIVCPSCTRR91 pKa = 11.84QNLQNGRR98 pKa = 11.84SHH100 pKa = 7.1
Molecular weight: 11.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G5ELP3|G5ELP3_9PAPI Uncharacterized protein HpV126gp5 OS=Human papillomavirus 126 OX=1055684 GN=HpV126gp5 PE=4 SV=1
MM1 pKa = 6.96NQNDD5 pKa = 3.41LRR7 pKa = 11.84EE8 pKa = 4.3RR9 pKa = 11.84LDD11 pKa = 3.7ALQTEE16 pKa = 5.01LMTLYY21 pKa = 7.84EE22 pKa = 4.4TNPTDD27 pKa = 3.76LKK29 pKa = 11.26SQITHH34 pKa = 5.25YY35 pKa = 10.8QLLRR39 pKa = 11.84KK40 pKa = 9.8EE41 pKa = 4.97GVLQYY46 pKa = 7.46YY47 pKa = 7.12TRR49 pKa = 11.84KK50 pKa = 9.97EE51 pKa = 4.18GYY53 pKa = 9.85DD54 pKa = 3.44RR55 pKa = 11.84LGLQYY60 pKa = 10.38IPPLKK65 pKa = 9.96ISEE68 pKa = 4.24NHH70 pKa = 6.13AKK72 pKa = 8.96TAIKK76 pKa = 9.52MILILEE82 pKa = 4.23SLAKK86 pKa = 9.94SKK88 pKa = 10.7YY89 pKa = 10.37ASEE92 pKa = 3.73QWTFADD98 pKa = 4.58CSADD102 pKa = 3.49RR103 pKa = 11.84FNSPPRR109 pKa = 11.84NCLKK113 pKa = 10.23KK114 pKa = 9.9HH115 pKa = 4.64SFEE118 pKa = 4.29VEE120 pKa = 3.53VWFDD124 pKa = 3.29SDD126 pKa = 3.38KK127 pKa = 11.53QNAFPYY133 pKa = 10.33INWRR137 pKa = 11.84DD138 pKa = 3.16IYY140 pKa = 11.15YY141 pKa = 10.02QDD143 pKa = 5.67EE144 pKa = 3.98KK145 pKa = 11.7DD146 pKa = 3.17EE147 pKa = 3.96WHH149 pKa = 6.16KK150 pKa = 11.56VKK152 pKa = 11.28GEE154 pKa = 3.58VDD156 pKa = 3.54YY157 pKa = 11.84NGLFFTEE164 pKa = 4.43LDD166 pKa = 3.21GTRR169 pKa = 11.84NYY171 pKa = 10.64FILFEE176 pKa = 5.48KK177 pKa = 10.84DD178 pKa = 2.36AFRR181 pKa = 11.84YY182 pKa = 9.49GKK184 pKa = 8.73TGTWTVNYY192 pKa = 9.87DD193 pKa = 3.39NEE195 pKa = 4.46QILPFVTSSSRR206 pKa = 11.84KK207 pKa = 9.7SISDD211 pKa = 3.46TQEE214 pKa = 3.57DD215 pKa = 4.82TTTGEE220 pKa = 4.3PSTSLSTSKK229 pKa = 10.89EE230 pKa = 3.4KK231 pKa = 10.79DD232 pKa = 3.02NRR234 pKa = 11.84RR235 pKa = 11.84RR236 pKa = 11.84TPGSQEE242 pKa = 3.86GSPSSTTRR250 pKa = 11.84HH251 pKa = 4.04GRR253 pKa = 11.84RR254 pKa = 11.84RR255 pKa = 11.84RR256 pKa = 11.84RR257 pKa = 11.84GGEE260 pKa = 3.68QRR262 pKa = 11.84EE263 pKa = 4.17SPPRR267 pKa = 11.84NKK269 pKa = 9.61RR270 pKa = 11.84RR271 pKa = 11.84RR272 pKa = 11.84GAQTDD277 pKa = 3.52GTADD281 pKa = 3.7YY282 pKa = 10.48ISPEE286 pKa = 4.16EE287 pKa = 3.91VGGSHH292 pKa = 7.4RR293 pKa = 11.84SVQRR297 pKa = 11.84TGLGRR302 pKa = 11.84IEE304 pKa = 4.07RR305 pKa = 11.84LKK307 pKa = 11.09EE308 pKa = 3.65EE309 pKa = 4.63ARR311 pKa = 11.84DD312 pKa = 3.67PPIIQLKK319 pKa = 10.42GGANSLKK326 pKa = 10.07CWRR329 pKa = 11.84NRR331 pKa = 11.84FKK333 pKa = 10.66IKK335 pKa = 10.91YY336 pKa = 8.53KK337 pKa = 10.9DD338 pKa = 3.6LFTAITTVYY347 pKa = 10.24KK348 pKa = 10.25WVGNNNADD356 pKa = 3.73EE357 pKa = 4.43VEE359 pKa = 4.2SRR361 pKa = 11.84LMVAFRR367 pKa = 11.84DD368 pKa = 3.79LSQRR372 pKa = 11.84SAFIATVHH380 pKa = 5.85IPKK383 pKa = 9.9GVSMSLGSLDD393 pKa = 3.83SLL395 pKa = 4.57
MM1 pKa = 6.96NQNDD5 pKa = 3.41LRR7 pKa = 11.84EE8 pKa = 4.3RR9 pKa = 11.84LDD11 pKa = 3.7ALQTEE16 pKa = 5.01LMTLYY21 pKa = 7.84EE22 pKa = 4.4TNPTDD27 pKa = 3.76LKK29 pKa = 11.26SQITHH34 pKa = 5.25YY35 pKa = 10.8QLLRR39 pKa = 11.84KK40 pKa = 9.8EE41 pKa = 4.97GVLQYY46 pKa = 7.46YY47 pKa = 7.12TRR49 pKa = 11.84KK50 pKa = 9.97EE51 pKa = 4.18GYY53 pKa = 9.85DD54 pKa = 3.44RR55 pKa = 11.84LGLQYY60 pKa = 10.38IPPLKK65 pKa = 9.96ISEE68 pKa = 4.24NHH70 pKa = 6.13AKK72 pKa = 8.96TAIKK76 pKa = 9.52MILILEE82 pKa = 4.23SLAKK86 pKa = 9.94SKK88 pKa = 10.7YY89 pKa = 10.37ASEE92 pKa = 3.73QWTFADD98 pKa = 4.58CSADD102 pKa = 3.49RR103 pKa = 11.84FNSPPRR109 pKa = 11.84NCLKK113 pKa = 10.23KK114 pKa = 9.9HH115 pKa = 4.64SFEE118 pKa = 4.29VEE120 pKa = 3.53VWFDD124 pKa = 3.29SDD126 pKa = 3.38KK127 pKa = 11.53QNAFPYY133 pKa = 10.33INWRR137 pKa = 11.84DD138 pKa = 3.16IYY140 pKa = 11.15YY141 pKa = 10.02QDD143 pKa = 5.67EE144 pKa = 3.98KK145 pKa = 11.7DD146 pKa = 3.17EE147 pKa = 3.96WHH149 pKa = 6.16KK150 pKa = 11.56VKK152 pKa = 11.28GEE154 pKa = 3.58VDD156 pKa = 3.54YY157 pKa = 11.84NGLFFTEE164 pKa = 4.43LDD166 pKa = 3.21GTRR169 pKa = 11.84NYY171 pKa = 10.64FILFEE176 pKa = 5.48KK177 pKa = 10.84DD178 pKa = 2.36AFRR181 pKa = 11.84YY182 pKa = 9.49GKK184 pKa = 8.73TGTWTVNYY192 pKa = 9.87DD193 pKa = 3.39NEE195 pKa = 4.46QILPFVTSSSRR206 pKa = 11.84KK207 pKa = 9.7SISDD211 pKa = 3.46TQEE214 pKa = 3.57DD215 pKa = 4.82TTTGEE220 pKa = 4.3PSTSLSTSKK229 pKa = 10.89EE230 pKa = 3.4KK231 pKa = 10.79DD232 pKa = 3.02NRR234 pKa = 11.84RR235 pKa = 11.84RR236 pKa = 11.84TPGSQEE242 pKa = 3.86GSPSSTTRR250 pKa = 11.84HH251 pKa = 4.04GRR253 pKa = 11.84RR254 pKa = 11.84RR255 pKa = 11.84RR256 pKa = 11.84RR257 pKa = 11.84GGEE260 pKa = 3.68QRR262 pKa = 11.84EE263 pKa = 4.17SPPRR267 pKa = 11.84NKK269 pKa = 9.61RR270 pKa = 11.84RR271 pKa = 11.84RR272 pKa = 11.84GAQTDD277 pKa = 3.52GTADD281 pKa = 3.7YY282 pKa = 10.48ISPEE286 pKa = 4.16EE287 pKa = 3.91VGGSHH292 pKa = 7.4RR293 pKa = 11.84SVQRR297 pKa = 11.84TGLGRR302 pKa = 11.84IEE304 pKa = 4.07RR305 pKa = 11.84LKK307 pKa = 11.09EE308 pKa = 3.65EE309 pKa = 4.63ARR311 pKa = 11.84DD312 pKa = 3.67PPIIQLKK319 pKa = 10.42GGANSLKK326 pKa = 10.07CWRR329 pKa = 11.84NRR331 pKa = 11.84FKK333 pKa = 10.66IKK335 pKa = 10.91YY336 pKa = 8.53KK337 pKa = 10.9DD338 pKa = 3.6LFTAITTVYY347 pKa = 10.24KK348 pKa = 10.25WVGNNNADD356 pKa = 3.73EE357 pKa = 4.43VEE359 pKa = 4.2SRR361 pKa = 11.84LMVAFRR367 pKa = 11.84DD368 pKa = 3.79LSQRR372 pKa = 11.84SAFIATVHH380 pKa = 5.85IPKK383 pKa = 9.9GVSMSLGSLDD393 pKa = 3.83SLL395 pKa = 4.57
Molecular weight: 45.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2439 |
100 |
607 |
348.4 |
39.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.453 ± 0.502 | 2.501 ± 0.905 |
6.56 ± 0.267 | 6.765 ± 0.598 |
4.715 ± 0.586 | 5.248 ± 0.763 |
1.804 ± 0.283 | 5.863 ± 0.53 |
5.781 ± 1.004 | 9.594 ± 1.127 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.763 ± 0.305 | 4.92 ± 0.426 |
6.15 ± 0.948 | 3.608 ± 0.247 |
5.904 ± 0.768 | 7.462 ± 0.636 |
6.396 ± 0.763 | 5.248 ± 0.435 |
1.23 ± 0.251 | 3.034 ± 0.301 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
