
Periconia macrospinosa
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Pleosporomycetidae; Pleosporales; Massarineae; Periconiaceae; Periconia
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
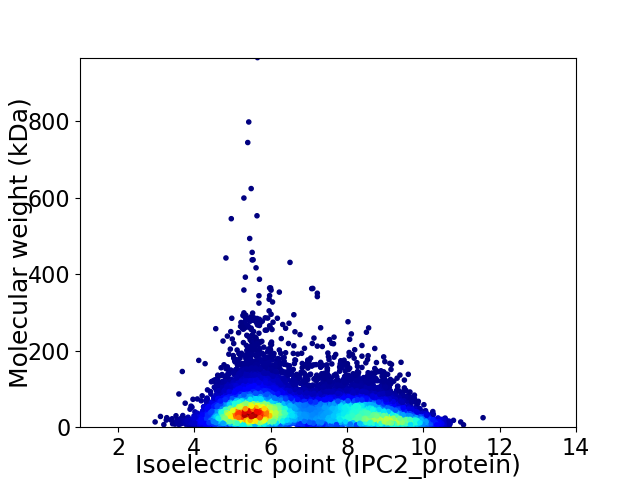
Virtual 2D-PAGE plot for 18722 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
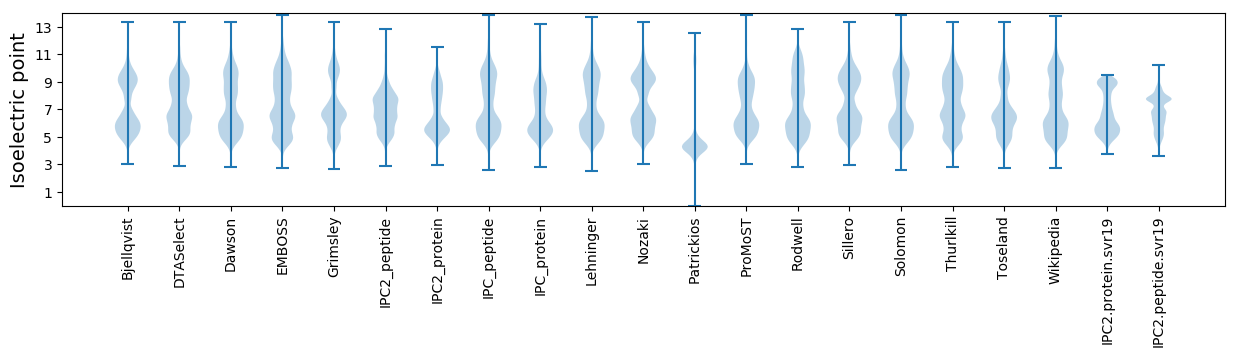
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2V1E641|A0A2V1E641_9PLEO V-type proton ATPase subunit a OS=Periconia macrospinosa OX=97972 GN=DM02DRAFT_725363 PE=3 SV=1
MM1 pKa = 7.0TTTAWATPGFEE12 pKa = 5.19LNPLSDD18 pKa = 3.56VMDD21 pKa = 3.47IHH23 pKa = 7.61SDD25 pKa = 3.25RR26 pKa = 11.84GVDD29 pKa = 3.52VDD31 pKa = 5.08NVDD34 pKa = 2.94VDD36 pKa = 3.56IDD38 pKa = 4.19FDD40 pKa = 4.26PSRR43 pKa = 11.84SPATYY48 pKa = 10.91GDD50 pKa = 4.13DD51 pKa = 5.17LSLKK55 pKa = 10.44DD56 pKa = 4.06AVLEE60 pKa = 4.29GEE62 pKa = 5.12PGPQTASADD71 pKa = 3.49QDD73 pKa = 3.72DD74 pKa = 4.31FMVDD78 pKa = 3.28NEE80 pKa = 4.55DD81 pKa = 4.97LIEE84 pKa = 4.61EE85 pKa = 4.05DD86 pKa = 3.58TIAYY90 pKa = 9.82EE91 pKa = 3.98EE92 pKa = 4.6DD93 pKa = 3.28AVIVDD98 pKa = 4.41EE99 pKa = 5.07PSANQNEE106 pKa = 4.39VDD108 pKa = 4.29HH109 pKa = 7.1SVDD112 pKa = 4.86DD113 pKa = 4.69DD114 pKa = 5.1LIDD117 pKa = 4.0YY118 pKa = 11.06SDD120 pKa = 5.09DD121 pKa = 4.25EE122 pKa = 4.59EE123 pKa = 6.69DD124 pKa = 3.67STPKK128 pKa = 10.52EE129 pKa = 4.03DD130 pKa = 5.09GISKK134 pKa = 8.5NTDD137 pKa = 2.69LTSARR142 pKa = 11.84NDD144 pKa = 3.3TAASVAEE151 pKa = 4.3SGPTEE156 pKa = 4.07IGPTSTEE163 pKa = 3.9IEE165 pKa = 4.27HH166 pKa = 6.15TAVDD170 pKa = 3.74PADD173 pKa = 4.34EE174 pKa = 4.23YY175 pKa = 11.72DD176 pKa = 3.41EE177 pKa = 5.17SKK179 pKa = 10.38IDD181 pKa = 3.65EE182 pKa = 4.88LEE184 pKa = 4.29TTGEE188 pKa = 4.13PGEE191 pKa = 4.22NWQEE195 pKa = 3.9AQEE198 pKa = 4.32AAHH201 pKa = 5.46QQEE204 pKa = 4.17LSIYY208 pKa = 10.76ADD210 pKa = 3.25VDD212 pKa = 4.3SNDD215 pKa = 3.6AQPEE219 pKa = 4.13EE220 pKa = 4.21NEE222 pKa = 4.31YY223 pKa = 11.31DD224 pKa = 4.15HH225 pKa = 7.39EE226 pKa = 4.51DD227 pKa = 3.75QGAQKK232 pKa = 10.26QDD234 pKa = 2.94NSEE237 pKa = 3.98DD238 pKa = 3.66VEE240 pKa = 4.78YY241 pKa = 10.81VAHH244 pKa = 6.98ASNDD248 pKa = 2.81IHH250 pKa = 7.55SPSGQEE256 pKa = 3.84IEE258 pKa = 4.29LHH260 pKa = 5.92PVTINYY266 pKa = 10.0SGAEE270 pKa = 3.58LWLFKK275 pKa = 10.79HH276 pKa = 6.55HH277 pKa = 7.5DD278 pKa = 3.88YY279 pKa = 11.27EE280 pKa = 6.28DD281 pKa = 3.68SGDD284 pKa = 3.92YY285 pKa = 10.64LVEE288 pKa = 4.37DD289 pKa = 3.85TSITNKK295 pKa = 9.97PISSVLEE302 pKa = 3.86ACRR305 pKa = 11.84SALGEE310 pKa = 4.46DD311 pKa = 3.4VTDD314 pKa = 5.09DD315 pKa = 3.43IEE317 pKa = 5.6LGFRR321 pKa = 11.84LDD323 pKa = 3.26NFRR326 pKa = 11.84NIEE329 pKa = 4.13LYY331 pKa = 10.09QDD333 pKa = 3.54HH334 pKa = 6.98SSCAFITLEE343 pKa = 4.64HH344 pKa = 6.42IVGLYY349 pKa = 9.85LQLHH353 pKa = 5.62AQDD356 pKa = 5.29GISDD360 pKa = 4.01PEE362 pKa = 4.21SFYY365 pKa = 9.02MTLLSRR371 pKa = 11.84PRR373 pKa = 11.84VSALFNALNEE383 pKa = 4.24AASEE387 pKa = 4.65GIGHH391 pKa = 6.48VGLEE395 pKa = 3.94KK396 pKa = 10.63AIAAGLTSFNARR408 pKa = 11.84LSHH411 pKa = 5.07NTAVNSYY418 pKa = 10.09DD419 pKa = 3.01WGNEE423 pKa = 3.78EE424 pKa = 3.97EE425 pKa = 4.59HH426 pKa = 6.27QGHH429 pKa = 5.87VADD432 pKa = 4.25VANGNQVEE440 pKa = 4.52GNGNGEE446 pKa = 4.18HH447 pKa = 5.76EE448 pKa = 4.45THH450 pKa = 6.47NEE452 pKa = 3.85HH453 pKa = 7.08EE454 pKa = 4.38EE455 pKa = 3.99EE456 pKa = 4.03EE457 pKa = 4.36QYY459 pKa = 11.44EE460 pKa = 4.39NSAQTHH466 pKa = 5.71EE467 pKa = 4.84GGAQEE472 pKa = 4.34SEE474 pKa = 4.14LVEE477 pKa = 4.66EE478 pKa = 5.05IIEE481 pKa = 4.41PTTQDD486 pKa = 3.21QNSFATDD493 pKa = 3.51GALSEE498 pKa = 4.63PEE500 pKa = 4.1NVTLNVADD508 pKa = 4.62NDD510 pKa = 4.7DD511 pKa = 4.1IQPAPATGSPRR522 pKa = 11.84TEE524 pKa = 3.94EE525 pKa = 3.72QAGDD529 pKa = 3.55NDD531 pKa = 4.12VQKK534 pKa = 11.06SPSQQEE540 pKa = 3.78DD541 pKa = 3.67DD542 pKa = 5.07DD543 pKa = 4.51FVDD546 pKa = 3.85YY547 pKa = 11.39SDD549 pKa = 6.2DD550 pKa = 3.74EE551 pKa = 5.6DD552 pKa = 3.84EE553 pKa = 6.06GEE555 pKa = 4.33PQPEE559 pKa = 3.95DD560 pKa = 3.57LARR563 pKa = 11.84QPSSLSSTVQGDD575 pKa = 3.44DD576 pKa = 3.38TFPVQEE582 pKa = 4.51IDD584 pKa = 4.07QEE586 pKa = 4.72TEE588 pKa = 3.71GLAVAEE594 pKa = 4.65HH595 pKa = 7.21DD596 pKa = 3.94PEE598 pKa = 4.18QSNDD602 pKa = 3.18YY603 pKa = 10.27HH604 pKa = 7.59VVVDD608 pKa = 4.06EE609 pKa = 4.47EE610 pKa = 5.94DD611 pKa = 3.5YY612 pKa = 11.46AQYY615 pKa = 11.7ADD617 pKa = 3.87ADD619 pKa = 3.93GGFDD623 pKa = 3.39AEE625 pKa = 5.45AYY627 pKa = 8.07EE628 pKa = 4.92QYY630 pKa = 10.99DD631 pKa = 3.46EE632 pKa = 5.99QYY634 pKa = 11.51DD635 pKa = 3.74EE636 pKa = 4.64LQAPEE641 pKa = 4.12HH642 pKa = 6.29TEE644 pKa = 3.76NDD646 pKa = 3.65DD647 pKa = 3.53AQEE650 pKa = 4.24AFQSQSYY657 pKa = 10.61EE658 pKa = 4.04DD659 pKa = 5.39LDD661 pKa = 4.11TNNYY665 pKa = 9.45NVGSEE670 pKa = 4.1EE671 pKa = 4.15QATGHH676 pKa = 6.29YY677 pKa = 9.68DD678 pKa = 3.22GKK680 pKa = 9.08TAGVDD685 pKa = 3.66GNYY688 pKa = 8.54EE689 pKa = 3.95AQVTVAGGGSIVGDD703 pKa = 3.72DD704 pKa = 3.55EE705 pKa = 6.21LEE707 pKa = 4.3NFDD710 pKa = 5.56LADD713 pKa = 3.43ATADD717 pKa = 3.51VTSYY721 pKa = 11.56DD722 pKa = 3.57AVGDD726 pKa = 3.67NAFNDD731 pKa = 4.02QYY733 pKa = 11.76SHH735 pKa = 6.94NDD737 pKa = 3.25EE738 pKa = 4.49NDD740 pKa = 3.2GDD742 pKa = 4.16GQAQVAAVSADD753 pKa = 3.72VEE755 pKa = 4.45PVAALSSEE763 pKa = 4.18SLNLSPQGQKK773 pKa = 8.78RR774 pKa = 11.84TIDD777 pKa = 3.43EE778 pKa = 4.17VGNDD782 pKa = 3.22VGEE785 pKa = 4.39ATDD788 pKa = 3.92PSDD791 pKa = 3.48AKK793 pKa = 10.52RR794 pKa = 11.84PRR796 pKa = 11.84VV797 pKa = 3.36
MM1 pKa = 7.0TTTAWATPGFEE12 pKa = 5.19LNPLSDD18 pKa = 3.56VMDD21 pKa = 3.47IHH23 pKa = 7.61SDD25 pKa = 3.25RR26 pKa = 11.84GVDD29 pKa = 3.52VDD31 pKa = 5.08NVDD34 pKa = 2.94VDD36 pKa = 3.56IDD38 pKa = 4.19FDD40 pKa = 4.26PSRR43 pKa = 11.84SPATYY48 pKa = 10.91GDD50 pKa = 4.13DD51 pKa = 5.17LSLKK55 pKa = 10.44DD56 pKa = 4.06AVLEE60 pKa = 4.29GEE62 pKa = 5.12PGPQTASADD71 pKa = 3.49QDD73 pKa = 3.72DD74 pKa = 4.31FMVDD78 pKa = 3.28NEE80 pKa = 4.55DD81 pKa = 4.97LIEE84 pKa = 4.61EE85 pKa = 4.05DD86 pKa = 3.58TIAYY90 pKa = 9.82EE91 pKa = 3.98EE92 pKa = 4.6DD93 pKa = 3.28AVIVDD98 pKa = 4.41EE99 pKa = 5.07PSANQNEE106 pKa = 4.39VDD108 pKa = 4.29HH109 pKa = 7.1SVDD112 pKa = 4.86DD113 pKa = 4.69DD114 pKa = 5.1LIDD117 pKa = 4.0YY118 pKa = 11.06SDD120 pKa = 5.09DD121 pKa = 4.25EE122 pKa = 4.59EE123 pKa = 6.69DD124 pKa = 3.67STPKK128 pKa = 10.52EE129 pKa = 4.03DD130 pKa = 5.09GISKK134 pKa = 8.5NTDD137 pKa = 2.69LTSARR142 pKa = 11.84NDD144 pKa = 3.3TAASVAEE151 pKa = 4.3SGPTEE156 pKa = 4.07IGPTSTEE163 pKa = 3.9IEE165 pKa = 4.27HH166 pKa = 6.15TAVDD170 pKa = 3.74PADD173 pKa = 4.34EE174 pKa = 4.23YY175 pKa = 11.72DD176 pKa = 3.41EE177 pKa = 5.17SKK179 pKa = 10.38IDD181 pKa = 3.65EE182 pKa = 4.88LEE184 pKa = 4.29TTGEE188 pKa = 4.13PGEE191 pKa = 4.22NWQEE195 pKa = 3.9AQEE198 pKa = 4.32AAHH201 pKa = 5.46QQEE204 pKa = 4.17LSIYY208 pKa = 10.76ADD210 pKa = 3.25VDD212 pKa = 4.3SNDD215 pKa = 3.6AQPEE219 pKa = 4.13EE220 pKa = 4.21NEE222 pKa = 4.31YY223 pKa = 11.31DD224 pKa = 4.15HH225 pKa = 7.39EE226 pKa = 4.51DD227 pKa = 3.75QGAQKK232 pKa = 10.26QDD234 pKa = 2.94NSEE237 pKa = 3.98DD238 pKa = 3.66VEE240 pKa = 4.78YY241 pKa = 10.81VAHH244 pKa = 6.98ASNDD248 pKa = 2.81IHH250 pKa = 7.55SPSGQEE256 pKa = 3.84IEE258 pKa = 4.29LHH260 pKa = 5.92PVTINYY266 pKa = 10.0SGAEE270 pKa = 3.58LWLFKK275 pKa = 10.79HH276 pKa = 6.55HH277 pKa = 7.5DD278 pKa = 3.88YY279 pKa = 11.27EE280 pKa = 6.28DD281 pKa = 3.68SGDD284 pKa = 3.92YY285 pKa = 10.64LVEE288 pKa = 4.37DD289 pKa = 3.85TSITNKK295 pKa = 9.97PISSVLEE302 pKa = 3.86ACRR305 pKa = 11.84SALGEE310 pKa = 4.46DD311 pKa = 3.4VTDD314 pKa = 5.09DD315 pKa = 3.43IEE317 pKa = 5.6LGFRR321 pKa = 11.84LDD323 pKa = 3.26NFRR326 pKa = 11.84NIEE329 pKa = 4.13LYY331 pKa = 10.09QDD333 pKa = 3.54HH334 pKa = 6.98SSCAFITLEE343 pKa = 4.64HH344 pKa = 6.42IVGLYY349 pKa = 9.85LQLHH353 pKa = 5.62AQDD356 pKa = 5.29GISDD360 pKa = 4.01PEE362 pKa = 4.21SFYY365 pKa = 9.02MTLLSRR371 pKa = 11.84PRR373 pKa = 11.84VSALFNALNEE383 pKa = 4.24AASEE387 pKa = 4.65GIGHH391 pKa = 6.48VGLEE395 pKa = 3.94KK396 pKa = 10.63AIAAGLTSFNARR408 pKa = 11.84LSHH411 pKa = 5.07NTAVNSYY418 pKa = 10.09DD419 pKa = 3.01WGNEE423 pKa = 3.78EE424 pKa = 3.97EE425 pKa = 4.59HH426 pKa = 6.27QGHH429 pKa = 5.87VADD432 pKa = 4.25VANGNQVEE440 pKa = 4.52GNGNGEE446 pKa = 4.18HH447 pKa = 5.76EE448 pKa = 4.45THH450 pKa = 6.47NEE452 pKa = 3.85HH453 pKa = 7.08EE454 pKa = 4.38EE455 pKa = 3.99EE456 pKa = 4.03EE457 pKa = 4.36QYY459 pKa = 11.44EE460 pKa = 4.39NSAQTHH466 pKa = 5.71EE467 pKa = 4.84GGAQEE472 pKa = 4.34SEE474 pKa = 4.14LVEE477 pKa = 4.66EE478 pKa = 5.05IIEE481 pKa = 4.41PTTQDD486 pKa = 3.21QNSFATDD493 pKa = 3.51GALSEE498 pKa = 4.63PEE500 pKa = 4.1NVTLNVADD508 pKa = 4.62NDD510 pKa = 4.7DD511 pKa = 4.1IQPAPATGSPRR522 pKa = 11.84TEE524 pKa = 3.94EE525 pKa = 3.72QAGDD529 pKa = 3.55NDD531 pKa = 4.12VQKK534 pKa = 11.06SPSQQEE540 pKa = 3.78DD541 pKa = 3.67DD542 pKa = 5.07DD543 pKa = 4.51FVDD546 pKa = 3.85YY547 pKa = 11.39SDD549 pKa = 6.2DD550 pKa = 3.74EE551 pKa = 5.6DD552 pKa = 3.84EE553 pKa = 6.06GEE555 pKa = 4.33PQPEE559 pKa = 3.95DD560 pKa = 3.57LARR563 pKa = 11.84QPSSLSSTVQGDD575 pKa = 3.44DD576 pKa = 3.38TFPVQEE582 pKa = 4.51IDD584 pKa = 4.07QEE586 pKa = 4.72TEE588 pKa = 3.71GLAVAEE594 pKa = 4.65HH595 pKa = 7.21DD596 pKa = 3.94PEE598 pKa = 4.18QSNDD602 pKa = 3.18YY603 pKa = 10.27HH604 pKa = 7.59VVVDD608 pKa = 4.06EE609 pKa = 4.47EE610 pKa = 5.94DD611 pKa = 3.5YY612 pKa = 11.46AQYY615 pKa = 11.7ADD617 pKa = 3.87ADD619 pKa = 3.93GGFDD623 pKa = 3.39AEE625 pKa = 5.45AYY627 pKa = 8.07EE628 pKa = 4.92QYY630 pKa = 10.99DD631 pKa = 3.46EE632 pKa = 5.99QYY634 pKa = 11.51DD635 pKa = 3.74EE636 pKa = 4.64LQAPEE641 pKa = 4.12HH642 pKa = 6.29TEE644 pKa = 3.76NDD646 pKa = 3.65DD647 pKa = 3.53AQEE650 pKa = 4.24AFQSQSYY657 pKa = 10.61EE658 pKa = 4.04DD659 pKa = 5.39LDD661 pKa = 4.11TNNYY665 pKa = 9.45NVGSEE670 pKa = 4.1EE671 pKa = 4.15QATGHH676 pKa = 6.29YY677 pKa = 9.68DD678 pKa = 3.22GKK680 pKa = 9.08TAGVDD685 pKa = 3.66GNYY688 pKa = 8.54EE689 pKa = 3.95AQVTVAGGGSIVGDD703 pKa = 3.72DD704 pKa = 3.55EE705 pKa = 6.21LEE707 pKa = 4.3NFDD710 pKa = 5.56LADD713 pKa = 3.43ATADD717 pKa = 3.51VTSYY721 pKa = 11.56DD722 pKa = 3.57AVGDD726 pKa = 3.67NAFNDD731 pKa = 4.02QYY733 pKa = 11.76SHH735 pKa = 6.94NDD737 pKa = 3.25EE738 pKa = 4.49NDD740 pKa = 3.2GDD742 pKa = 4.16GQAQVAAVSADD753 pKa = 3.72VEE755 pKa = 4.45PVAALSSEE763 pKa = 4.18SLNLSPQGQKK773 pKa = 8.78RR774 pKa = 11.84TIDD777 pKa = 3.43EE778 pKa = 4.17VGNDD782 pKa = 3.22VGEE785 pKa = 4.39ATDD788 pKa = 3.92PSDD791 pKa = 3.48AKK793 pKa = 10.52RR794 pKa = 11.84PRR796 pKa = 11.84VV797 pKa = 3.36
Molecular weight: 86.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2V1DCG3|A0A2V1DCG3_9PLEO Uncharacterized protein OS=Periconia macrospinosa OX=97972 GN=DM02DRAFT_138206 PE=4 SV=1
MM1 pKa = 7.39ILSNRR6 pKa = 11.84RR7 pKa = 11.84PQPNQNPLHH16 pKa = 7.04RR17 pKa = 11.84IQLQNLPQNPLRR29 pKa = 11.84HH30 pKa = 5.04QLKK33 pKa = 8.92MLRR36 pKa = 11.84QIQPRR41 pKa = 11.84HH42 pKa = 5.54KK43 pKa = 10.24RR44 pKa = 11.84RR45 pKa = 11.84NLPSRR50 pKa = 11.84QRR52 pKa = 11.84RR53 pKa = 11.84SILPRR58 pKa = 11.84APEE61 pKa = 4.03TLPPNHH67 pKa = 6.91LLHH70 pKa = 6.34RR71 pKa = 11.84QNRR74 pKa = 11.84PPRR77 pKa = 11.84LGNHH81 pKa = 6.66PPRR84 pKa = 11.84QRR86 pKa = 11.84RR87 pKa = 11.84QRR89 pKa = 11.84QHH91 pKa = 6.94PLPLPNLLLRR101 pKa = 11.84RR102 pKa = 11.84HH103 pKa = 6.24HH104 pKa = 6.52PVPVPLPQRR113 pKa = 11.84LRR115 pKa = 11.84APGGPLHH122 pKa = 6.28GALRR126 pKa = 11.84RR127 pKa = 11.84LHH129 pKa = 6.2RR130 pKa = 11.84RR131 pKa = 11.84RR132 pKa = 11.84SRR134 pKa = 11.84RR135 pKa = 11.84LGAPFIHH142 pKa = 6.72PHH144 pKa = 5.71HH145 pKa = 6.96RR146 pKa = 11.84PHH148 pKa = 7.52LGPRR152 pKa = 11.84PQRR155 pKa = 11.84PNPNRR160 pKa = 11.84PLPHH164 pKa = 7.09PSKK167 pKa = 10.72SPPRR171 pKa = 11.84RR172 pKa = 11.84QQRR175 pKa = 11.84RR176 pKa = 11.84HH177 pKa = 5.91LPFAPFPRR185 pKa = 11.84HH186 pKa = 5.27LPPRR190 pKa = 11.84RR191 pKa = 11.84LQILRR196 pKa = 11.84RR197 pKa = 11.84IHH199 pKa = 5.94HH200 pKa = 6.64LVRR203 pKa = 11.84ARR205 pKa = 11.84RR206 pKa = 3.48
MM1 pKa = 7.39ILSNRR6 pKa = 11.84RR7 pKa = 11.84PQPNQNPLHH16 pKa = 7.04RR17 pKa = 11.84IQLQNLPQNPLRR29 pKa = 11.84HH30 pKa = 5.04QLKK33 pKa = 8.92MLRR36 pKa = 11.84QIQPRR41 pKa = 11.84HH42 pKa = 5.54KK43 pKa = 10.24RR44 pKa = 11.84RR45 pKa = 11.84NLPSRR50 pKa = 11.84QRR52 pKa = 11.84RR53 pKa = 11.84SILPRR58 pKa = 11.84APEE61 pKa = 4.03TLPPNHH67 pKa = 6.91LLHH70 pKa = 6.34RR71 pKa = 11.84QNRR74 pKa = 11.84PPRR77 pKa = 11.84LGNHH81 pKa = 6.66PPRR84 pKa = 11.84QRR86 pKa = 11.84RR87 pKa = 11.84QRR89 pKa = 11.84QHH91 pKa = 6.94PLPLPNLLLRR101 pKa = 11.84RR102 pKa = 11.84HH103 pKa = 6.24HH104 pKa = 6.52PVPVPLPQRR113 pKa = 11.84LRR115 pKa = 11.84APGGPLHH122 pKa = 6.28GALRR126 pKa = 11.84RR127 pKa = 11.84LHH129 pKa = 6.2RR130 pKa = 11.84RR131 pKa = 11.84RR132 pKa = 11.84SRR134 pKa = 11.84RR135 pKa = 11.84LGAPFIHH142 pKa = 6.72PHH144 pKa = 5.71HH145 pKa = 6.96RR146 pKa = 11.84PHH148 pKa = 7.52LGPRR152 pKa = 11.84PQRR155 pKa = 11.84PNPNRR160 pKa = 11.84PLPHH164 pKa = 7.09PSKK167 pKa = 10.72SPPRR171 pKa = 11.84RR172 pKa = 11.84QQRR175 pKa = 11.84RR176 pKa = 11.84HH177 pKa = 5.91LPFAPFPRR185 pKa = 11.84HH186 pKa = 5.27LPPRR190 pKa = 11.84RR191 pKa = 11.84LQILRR196 pKa = 11.84RR197 pKa = 11.84IHH199 pKa = 5.94HH200 pKa = 6.64LVRR203 pKa = 11.84ARR205 pKa = 11.84RR206 pKa = 3.48
Molecular weight: 24.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7677607 |
49 |
8697 |
410.1 |
45.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.361 ± 0.016 | 1.364 ± 0.007 |
5.422 ± 0.014 | 6.025 ± 0.016 |
3.871 ± 0.01 | 6.687 ± 0.02 |
2.514 ± 0.008 | 5.098 ± 0.011 |
5.006 ± 0.014 | 8.936 ± 0.018 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.176 ± 0.007 | 3.868 ± 0.011 |
5.992 ± 0.018 | 3.954 ± 0.015 |
5.958 ± 0.015 | 8.138 ± 0.018 |
6.116 ± 0.013 | 6.071 ± 0.014 |
1.57 ± 0.007 | 2.872 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
