
Salanga virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Salanga phlebovirus
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
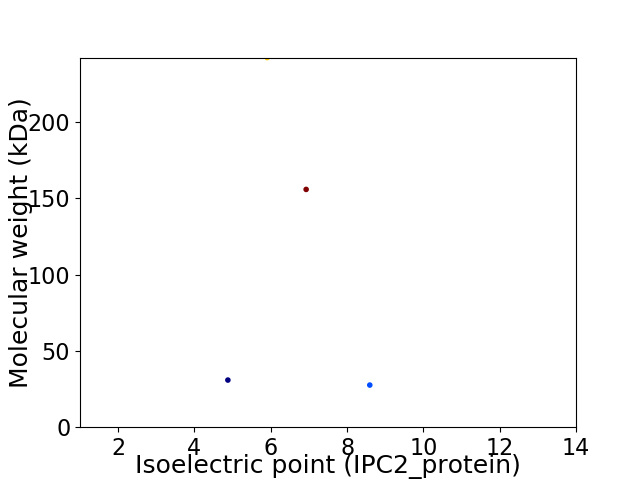
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U5XLH5|U5XLH5_9VIRU Replicase OS=Salanga virus OX=1416745 PE=4 SV=1
MM1 pKa = 8.01DD2 pKa = 5.81RR3 pKa = 11.84YY4 pKa = 10.07LHH6 pKa = 5.99NRR8 pKa = 11.84AIVHH12 pKa = 6.5RR13 pKa = 11.84LHH15 pKa = 6.91GSSNLPVVTFKK26 pKa = 11.2VIEE29 pKa = 4.31DD30 pKa = 5.08DD31 pKa = 3.34IDD33 pKa = 3.44EE34 pKa = 4.64HH35 pKa = 7.76VAVHH39 pKa = 5.13QCVEE43 pKa = 4.5VVLHH47 pKa = 4.95SHH49 pKa = 6.48RR50 pKa = 11.84LSRR53 pKa = 11.84AEE55 pKa = 3.81GHH57 pKa = 4.86SLKK60 pKa = 10.43WFLEE64 pKa = 4.02RR65 pKa = 11.84GLFPARR71 pKa = 11.84SGDD74 pKa = 4.47LIWSSCITRR83 pKa = 11.84IEE85 pKa = 4.13PQDD88 pKa = 3.91LNSYY92 pKa = 10.38LYY94 pKa = 11.03GFAALPRR101 pKa = 11.84EE102 pKa = 4.49DD103 pKa = 5.11FNCLQLRR110 pKa = 11.84NVRR113 pKa = 11.84KK114 pKa = 9.79ALEE117 pKa = 4.2WPTGQLSFEE126 pKa = 4.23FFDD129 pKa = 5.08LEE131 pKa = 4.56FPGKK135 pKa = 9.62YY136 pKa = 8.13VWAFEE141 pKa = 3.88NMRR144 pKa = 11.84RR145 pKa = 11.84TIQLLSMMSSSDD157 pKa = 3.33QVEE160 pKa = 5.18DD161 pKa = 4.81IICDD165 pKa = 3.37AYY167 pKa = 11.23DD168 pKa = 3.48CVLRR172 pKa = 11.84AGSDD176 pKa = 2.99LGLAHH181 pKa = 6.43STFPGNNIVYY191 pKa = 7.03EE192 pKa = 4.02VCYY195 pKa = 10.31IQLLKK200 pKa = 10.69AVQAYY205 pKa = 7.92STDD208 pKa = 3.22IQQVSGFPDD217 pKa = 3.14NAVTIFLKK225 pKa = 10.79DD226 pKa = 2.98IMPYY230 pKa = 10.08VRR232 pKa = 11.84DD233 pKa = 3.54RR234 pKa = 11.84FPGVRR239 pKa = 11.84EE240 pKa = 3.95PMSNPLPEE248 pKa = 4.84PSEE251 pKa = 4.02HH252 pKa = 6.39QLNVWALPDD261 pKa = 3.48EE262 pKa = 5.12GIEE265 pKa = 4.04EE266 pKa = 5.41DD267 pKa = 3.87IDD269 pKa = 3.56
MM1 pKa = 8.01DD2 pKa = 5.81RR3 pKa = 11.84YY4 pKa = 10.07LHH6 pKa = 5.99NRR8 pKa = 11.84AIVHH12 pKa = 6.5RR13 pKa = 11.84LHH15 pKa = 6.91GSSNLPVVTFKK26 pKa = 11.2VIEE29 pKa = 4.31DD30 pKa = 5.08DD31 pKa = 3.34IDD33 pKa = 3.44EE34 pKa = 4.64HH35 pKa = 7.76VAVHH39 pKa = 5.13QCVEE43 pKa = 4.5VVLHH47 pKa = 4.95SHH49 pKa = 6.48RR50 pKa = 11.84LSRR53 pKa = 11.84AEE55 pKa = 3.81GHH57 pKa = 4.86SLKK60 pKa = 10.43WFLEE64 pKa = 4.02RR65 pKa = 11.84GLFPARR71 pKa = 11.84SGDD74 pKa = 4.47LIWSSCITRR83 pKa = 11.84IEE85 pKa = 4.13PQDD88 pKa = 3.91LNSYY92 pKa = 10.38LYY94 pKa = 11.03GFAALPRR101 pKa = 11.84EE102 pKa = 4.49DD103 pKa = 5.11FNCLQLRR110 pKa = 11.84NVRR113 pKa = 11.84KK114 pKa = 9.79ALEE117 pKa = 4.2WPTGQLSFEE126 pKa = 4.23FFDD129 pKa = 5.08LEE131 pKa = 4.56FPGKK135 pKa = 9.62YY136 pKa = 8.13VWAFEE141 pKa = 3.88NMRR144 pKa = 11.84RR145 pKa = 11.84TIQLLSMMSSSDD157 pKa = 3.33QVEE160 pKa = 5.18DD161 pKa = 4.81IICDD165 pKa = 3.37AYY167 pKa = 11.23DD168 pKa = 3.48CVLRR172 pKa = 11.84AGSDD176 pKa = 2.99LGLAHH181 pKa = 6.43STFPGNNIVYY191 pKa = 7.03EE192 pKa = 4.02VCYY195 pKa = 10.31IQLLKK200 pKa = 10.69AVQAYY205 pKa = 7.92STDD208 pKa = 3.22IQQVSGFPDD217 pKa = 3.14NAVTIFLKK225 pKa = 10.79DD226 pKa = 2.98IMPYY230 pKa = 10.08VRR232 pKa = 11.84DD233 pKa = 3.54RR234 pKa = 11.84FPGVRR239 pKa = 11.84EE240 pKa = 3.95PMSNPLPEE248 pKa = 4.84PSEE251 pKa = 4.02HH252 pKa = 6.39QLNVWALPDD261 pKa = 3.48EE262 pKa = 5.12GIEE265 pKa = 4.04EE266 pKa = 5.41DD267 pKa = 3.87IDD269 pKa = 3.56
Molecular weight: 30.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U5XJM5|U5XJM5_9VIRU Nonstructural protein OS=Salanga virus OX=1416745 PE=4 SV=1
MM1 pKa = 8.27DD2 pKa = 4.04YY3 pKa = 11.53AKK5 pKa = 10.37IALDD9 pKa = 3.65FAGEE13 pKa = 4.53GISQVEE19 pKa = 3.82IAKK22 pKa = 9.16WVEE25 pKa = 3.56DD26 pKa = 3.74FAYY29 pKa = 10.39QGFDD33 pKa = 2.8AKK35 pKa = 10.47RR36 pKa = 11.84VVEE39 pKa = 4.1LVQTRR44 pKa = 11.84GSGRR48 pKa = 11.84NWKK51 pKa = 9.73QDD53 pKa = 3.35VAKK56 pKa = 9.71MIVIALTRR64 pKa = 11.84GNKK67 pKa = 7.49IAKK70 pKa = 7.94MKK72 pKa = 10.82QKK74 pKa = 10.2MSDD77 pKa = 3.12KK78 pKa = 10.45GKK80 pKa = 10.57EE81 pKa = 4.0SLTEE85 pKa = 3.82LEE87 pKa = 3.94RR88 pKa = 11.84VYY90 pKa = 10.13MLKK93 pKa = 10.38SGNPGRR99 pKa = 11.84DD100 pKa = 3.4DD101 pKa = 3.55LTLSRR106 pKa = 11.84IAVAFAPWTVGALNHH121 pKa = 6.99LDD123 pKa = 2.83GHH125 pKa = 6.53LPVNGKK131 pKa = 10.19DD132 pKa = 3.3MDD134 pKa = 3.93EE135 pKa = 4.2HH136 pKa = 5.37VQRR139 pKa = 11.84YY140 pKa = 6.1PRR142 pKa = 11.84CMMHH146 pKa = 6.86PAFGSLIDD154 pKa = 3.87RR155 pKa = 11.84NLPEE159 pKa = 4.12VTVSQLINAHH169 pKa = 6.72CIFLDD174 pKa = 3.26KK175 pKa = 10.05FARR178 pKa = 11.84VINPNLRR185 pKa = 11.84GKK187 pKa = 8.37TKK189 pKa = 10.52QEE191 pKa = 3.73VAKK194 pKa = 10.7SYY196 pKa = 7.4EE197 pKa = 4.18QPLKK201 pKa = 10.79AAVSSTFFSLEE212 pKa = 3.68QKK214 pKa = 10.61RR215 pKa = 11.84GVLKK219 pKa = 10.82NLGLIDD225 pKa = 4.25EE226 pKa = 4.51NLKK229 pKa = 8.52VTEE232 pKa = 4.23VVKK235 pKa = 10.47RR236 pKa = 11.84ASDD239 pKa = 3.75FWTQAFF245 pKa = 3.82
MM1 pKa = 8.27DD2 pKa = 4.04YY3 pKa = 11.53AKK5 pKa = 10.37IALDD9 pKa = 3.65FAGEE13 pKa = 4.53GISQVEE19 pKa = 3.82IAKK22 pKa = 9.16WVEE25 pKa = 3.56DD26 pKa = 3.74FAYY29 pKa = 10.39QGFDD33 pKa = 2.8AKK35 pKa = 10.47RR36 pKa = 11.84VVEE39 pKa = 4.1LVQTRR44 pKa = 11.84GSGRR48 pKa = 11.84NWKK51 pKa = 9.73QDD53 pKa = 3.35VAKK56 pKa = 9.71MIVIALTRR64 pKa = 11.84GNKK67 pKa = 7.49IAKK70 pKa = 7.94MKK72 pKa = 10.82QKK74 pKa = 10.2MSDD77 pKa = 3.12KK78 pKa = 10.45GKK80 pKa = 10.57EE81 pKa = 4.0SLTEE85 pKa = 3.82LEE87 pKa = 3.94RR88 pKa = 11.84VYY90 pKa = 10.13MLKK93 pKa = 10.38SGNPGRR99 pKa = 11.84DD100 pKa = 3.4DD101 pKa = 3.55LTLSRR106 pKa = 11.84IAVAFAPWTVGALNHH121 pKa = 6.99LDD123 pKa = 2.83GHH125 pKa = 6.53LPVNGKK131 pKa = 10.19DD132 pKa = 3.3MDD134 pKa = 3.93EE135 pKa = 4.2HH136 pKa = 5.37VQRR139 pKa = 11.84YY140 pKa = 6.1PRR142 pKa = 11.84CMMHH146 pKa = 6.86PAFGSLIDD154 pKa = 3.87RR155 pKa = 11.84NLPEE159 pKa = 4.12VTVSQLINAHH169 pKa = 6.72CIFLDD174 pKa = 3.26KK175 pKa = 10.05FARR178 pKa = 11.84VINPNLRR185 pKa = 11.84GKK187 pKa = 8.37TKK189 pKa = 10.52QEE191 pKa = 3.73VAKK194 pKa = 10.7SYY196 pKa = 7.4EE197 pKa = 4.18QPLKK201 pKa = 10.79AAVSSTFFSLEE212 pKa = 3.68QKK214 pKa = 10.61RR215 pKa = 11.84GVLKK219 pKa = 10.82NLGLIDD225 pKa = 4.25EE226 pKa = 4.51NLKK229 pKa = 8.52VTEE232 pKa = 4.23VVKK235 pKa = 10.47RR236 pKa = 11.84ASDD239 pKa = 3.75FWTQAFF245 pKa = 3.82
Molecular weight: 27.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4035 |
245 |
2123 |
1008.8 |
114.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.527 ± 0.414 | 2.677 ± 0.741 |
6.245 ± 0.351 | 6.295 ± 0.327 |
4.411 ± 0.133 | 5.651 ± 0.252 |
2.354 ± 0.151 | 6.072 ± 0.369 |
6.221 ± 0.367 | 9.021 ± 0.406 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.47 ± 0.572 | 4.04 ± 0.27 |
3.866 ± 0.175 | 3.371 ± 0.274 |
5.75 ± 0.167 | 8.848 ± 0.69 |
5.353 ± 0.461 | 6.667 ± 0.391 |
1.388 ± 0.115 | 2.776 ± 0.183 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
