
Halovenus aranensis
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Halobacteriaceae; Halovenus
Average proteome isoelectric point is 4.79
Get precalculated fractions of proteins
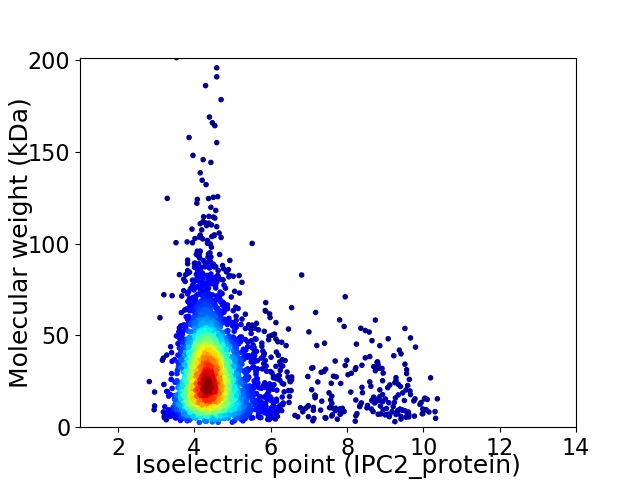
Virtual 2D-PAGE plot for 3354 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G8YLL0|A0A1G8YLL0_9EURY Dihydroneopterin aldolase OS=Halovenus aranensis OX=890420 GN=mptD PE=3 SV=1
MM1 pKa = 7.3NPEE4 pKa = 4.75DD5 pKa = 4.46FPTPAVDD12 pKa = 3.81VEE14 pKa = 4.89SVAPEE19 pKa = 4.04TLRR22 pKa = 11.84GRR24 pKa = 11.84IDD26 pKa = 3.24ANEE29 pKa = 3.97RR30 pKa = 11.84VTLLDD35 pKa = 3.24ARR37 pKa = 11.84MEE39 pKa = 4.17SEE41 pKa = 3.78YY42 pKa = 11.2DD43 pKa = 2.91EE44 pKa = 4.15WHH46 pKa = 6.46IDD48 pKa = 3.66GPSVEE53 pKa = 5.27SINIPYY59 pKa = 10.0FHH61 pKa = 7.18FLDD64 pKa = 5.52DD65 pKa = 4.36EE66 pKa = 4.69VDD68 pKa = 3.86DD69 pKa = 5.24DD70 pKa = 4.72VLAEE74 pKa = 4.16VPDD77 pKa = 3.97DD78 pKa = 4.15RR79 pKa = 11.84EE80 pKa = 4.37VTVLCAKK87 pKa = 10.37GGASEE92 pKa = 4.4YY93 pKa = 10.86VAGTLAEE100 pKa = 4.3LGYY103 pKa = 10.66EE104 pKa = 4.0VDD106 pKa = 3.77HH107 pKa = 8.28LEE109 pKa = 4.09EE110 pKa = 5.17GMNGWATIYY119 pKa = 10.74DD120 pKa = 3.73AVEE123 pKa = 3.74VTGYY127 pKa = 10.94DD128 pKa = 3.28GAGTLLQYY136 pKa = 10.06QRR138 pKa = 11.84PSSGCLGYY146 pKa = 11.15LLVDD150 pKa = 3.89GGEE153 pKa = 4.1AAVIDD158 pKa = 4.42PLRR161 pKa = 11.84AFTDD165 pKa = 4.04RR166 pKa = 11.84YY167 pKa = 10.49LADD170 pKa = 4.68ADD172 pKa = 4.52DD173 pKa = 5.2LGVDD177 pKa = 4.41LKK179 pKa = 11.32YY180 pKa = 11.02ALDD183 pKa = 3.64THH185 pKa = 6.8IHH187 pKa = 6.68ADD189 pKa = 3.7HH190 pKa = 6.89ISGVRR195 pKa = 11.84DD196 pKa = 3.4LDD198 pKa = 3.87AEE200 pKa = 4.39GVEE203 pKa = 4.6GVIPEE208 pKa = 4.09AAVDD212 pKa = 3.7RR213 pKa = 11.84GVTYY217 pKa = 10.86AEE219 pKa = 4.38EE220 pKa = 4.18LTTAADD226 pKa = 3.77GDD228 pKa = 4.45TIAVGDD234 pKa = 3.84ATIEE238 pKa = 4.31TVHH241 pKa = 6.22TPGHH245 pKa = 4.77TTGMTSYY252 pKa = 10.98LVDD255 pKa = 4.36DD256 pKa = 4.7SFLATGDD263 pKa = 3.76GLFVEE268 pKa = 5.56SVARR272 pKa = 11.84PDD274 pKa = 4.52LEE276 pKa = 4.65EE277 pKa = 6.06GDD279 pKa = 4.23EE280 pKa = 4.34GAPDD284 pKa = 4.07AARR287 pKa = 11.84MLYY290 pKa = 7.91EE291 pKa = 4.06TLQEE295 pKa = 4.78RR296 pKa = 11.84ILALPDD302 pKa = 3.1EE303 pKa = 4.93TIVGGAHH310 pKa = 7.0ASDD313 pKa = 3.58AAAAAEE319 pKa = 4.68DD320 pKa = 4.01GTYY323 pKa = 8.3TAPLGDD329 pKa = 4.85LVAEE333 pKa = 4.32MDD335 pKa = 4.27ALTMDD340 pKa = 3.9EE341 pKa = 4.43EE342 pKa = 4.73AFVDD346 pKa = 5.27LVLADD351 pKa = 3.95MPPRR355 pKa = 11.84PANYY359 pKa = 10.08EE360 pKa = 3.96EE361 pKa = 5.46IIATNLGQNSVDD373 pKa = 3.71DD374 pKa = 4.51EE375 pKa = 4.46EE376 pKa = 6.51AFTLEE381 pKa = 4.99LGPNNCAASQEE392 pKa = 4.43SLAGDD397 pKa = 3.77
MM1 pKa = 7.3NPEE4 pKa = 4.75DD5 pKa = 4.46FPTPAVDD12 pKa = 3.81VEE14 pKa = 4.89SVAPEE19 pKa = 4.04TLRR22 pKa = 11.84GRR24 pKa = 11.84IDD26 pKa = 3.24ANEE29 pKa = 3.97RR30 pKa = 11.84VTLLDD35 pKa = 3.24ARR37 pKa = 11.84MEE39 pKa = 4.17SEE41 pKa = 3.78YY42 pKa = 11.2DD43 pKa = 2.91EE44 pKa = 4.15WHH46 pKa = 6.46IDD48 pKa = 3.66GPSVEE53 pKa = 5.27SINIPYY59 pKa = 10.0FHH61 pKa = 7.18FLDD64 pKa = 5.52DD65 pKa = 4.36EE66 pKa = 4.69VDD68 pKa = 3.86DD69 pKa = 5.24DD70 pKa = 4.72VLAEE74 pKa = 4.16VPDD77 pKa = 3.97DD78 pKa = 4.15RR79 pKa = 11.84EE80 pKa = 4.37VTVLCAKK87 pKa = 10.37GGASEE92 pKa = 4.4YY93 pKa = 10.86VAGTLAEE100 pKa = 4.3LGYY103 pKa = 10.66EE104 pKa = 4.0VDD106 pKa = 3.77HH107 pKa = 8.28LEE109 pKa = 4.09EE110 pKa = 5.17GMNGWATIYY119 pKa = 10.74DD120 pKa = 3.73AVEE123 pKa = 3.74VTGYY127 pKa = 10.94DD128 pKa = 3.28GAGTLLQYY136 pKa = 10.06QRR138 pKa = 11.84PSSGCLGYY146 pKa = 11.15LLVDD150 pKa = 3.89GGEE153 pKa = 4.1AAVIDD158 pKa = 4.42PLRR161 pKa = 11.84AFTDD165 pKa = 4.04RR166 pKa = 11.84YY167 pKa = 10.49LADD170 pKa = 4.68ADD172 pKa = 4.52DD173 pKa = 5.2LGVDD177 pKa = 4.41LKK179 pKa = 11.32YY180 pKa = 11.02ALDD183 pKa = 3.64THH185 pKa = 6.8IHH187 pKa = 6.68ADD189 pKa = 3.7HH190 pKa = 6.89ISGVRR195 pKa = 11.84DD196 pKa = 3.4LDD198 pKa = 3.87AEE200 pKa = 4.39GVEE203 pKa = 4.6GVIPEE208 pKa = 4.09AAVDD212 pKa = 3.7RR213 pKa = 11.84GVTYY217 pKa = 10.86AEE219 pKa = 4.38EE220 pKa = 4.18LTTAADD226 pKa = 3.77GDD228 pKa = 4.45TIAVGDD234 pKa = 3.84ATIEE238 pKa = 4.31TVHH241 pKa = 6.22TPGHH245 pKa = 4.77TTGMTSYY252 pKa = 10.98LVDD255 pKa = 4.36DD256 pKa = 4.7SFLATGDD263 pKa = 3.76GLFVEE268 pKa = 5.56SVARR272 pKa = 11.84PDD274 pKa = 4.52LEE276 pKa = 4.65EE277 pKa = 6.06GDD279 pKa = 4.23EE280 pKa = 4.34GAPDD284 pKa = 4.07AARR287 pKa = 11.84MLYY290 pKa = 7.91EE291 pKa = 4.06TLQEE295 pKa = 4.78RR296 pKa = 11.84ILALPDD302 pKa = 3.1EE303 pKa = 4.93TIVGGAHH310 pKa = 7.0ASDD313 pKa = 3.58AAAAAEE319 pKa = 4.68DD320 pKa = 4.01GTYY323 pKa = 8.3TAPLGDD329 pKa = 4.85LVAEE333 pKa = 4.32MDD335 pKa = 4.27ALTMDD340 pKa = 3.9EE341 pKa = 4.43EE342 pKa = 4.73AFVDD346 pKa = 5.27LVLADD351 pKa = 3.95MPPRR355 pKa = 11.84PANYY359 pKa = 10.08EE360 pKa = 3.96EE361 pKa = 5.46IIATNLGQNSVDD373 pKa = 3.71DD374 pKa = 4.51EE375 pKa = 4.46EE376 pKa = 6.51AFTLEE381 pKa = 4.99LGPNNCAASQEE392 pKa = 4.43SLAGDD397 pKa = 3.77
Molecular weight: 42.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G8YV29|A0A1G8YV29_9EURY Protein-export membrane protein SecF OS=Halovenus aranensis OX=890420 GN=secF PE=3 SV=1
MM1 pKa = 7.28ARR3 pKa = 11.84SFYY6 pKa = 11.12SHH8 pKa = 7.07IRR10 pKa = 11.84DD11 pKa = 3.27AWKK14 pKa = 10.73DD15 pKa = 3.42PDD17 pKa = 4.17DD18 pKa = 4.52GKK20 pKa = 11.07VAQLQWEE27 pKa = 4.48RR28 pKa = 11.84QQEE31 pKa = 4.02WRR33 pKa = 11.84DD34 pKa = 3.22QGAIEE39 pKa = 4.73RR40 pKa = 11.84IEE42 pKa = 4.1RR43 pKa = 11.84PTRR46 pKa = 11.84LDD48 pKa = 3.12RR49 pKa = 11.84ARR51 pKa = 11.84SLGYY55 pKa = 9.43KK56 pKa = 9.87AKK58 pKa = 10.46QGVVLARR65 pKa = 11.84VSVRR69 pKa = 11.84KK70 pKa = 10.02GGSRR74 pKa = 11.84KK75 pKa = 8.19QRR77 pKa = 11.84FTAGRR82 pKa = 11.84RR83 pKa = 11.84SARR86 pKa = 11.84QGVNRR91 pKa = 11.84LNRR94 pKa = 11.84RR95 pKa = 11.84KK96 pKa = 9.97NIQRR100 pKa = 11.84IAEE103 pKa = 4.08EE104 pKa = 3.88RR105 pKa = 11.84CTRR108 pKa = 11.84KK109 pKa = 8.44FTNLRR114 pKa = 11.84VLNSYY119 pKa = 8.44WVGEE123 pKa = 4.4DD124 pKa = 4.78GSQKK128 pKa = 8.97WFEE131 pKa = 4.62VIMLDD136 pKa = 3.77PEE138 pKa = 4.41HH139 pKa = 6.93PAIEE143 pKa = 4.75NDD145 pKa = 5.08DD146 pKa = 4.12DD147 pKa = 5.67LNWICDD153 pKa = 3.7DD154 pKa = 3.61THH156 pKa = 7.42KK157 pKa = 10.93NRR159 pKa = 11.84AMRR162 pKa = 11.84GLTSAGTQNRR172 pKa = 11.84GLEE175 pKa = 4.37SKK177 pKa = 11.04GKK179 pKa = 8.02GTEE182 pKa = 3.7RR183 pKa = 11.84TRR185 pKa = 11.84PSLSSNKK192 pKa = 9.22SRR194 pKa = 11.84KK195 pKa = 8.98
MM1 pKa = 7.28ARR3 pKa = 11.84SFYY6 pKa = 11.12SHH8 pKa = 7.07IRR10 pKa = 11.84DD11 pKa = 3.27AWKK14 pKa = 10.73DD15 pKa = 3.42PDD17 pKa = 4.17DD18 pKa = 4.52GKK20 pKa = 11.07VAQLQWEE27 pKa = 4.48RR28 pKa = 11.84QQEE31 pKa = 4.02WRR33 pKa = 11.84DD34 pKa = 3.22QGAIEE39 pKa = 4.73RR40 pKa = 11.84IEE42 pKa = 4.1RR43 pKa = 11.84PTRR46 pKa = 11.84LDD48 pKa = 3.12RR49 pKa = 11.84ARR51 pKa = 11.84SLGYY55 pKa = 9.43KK56 pKa = 9.87AKK58 pKa = 10.46QGVVLARR65 pKa = 11.84VSVRR69 pKa = 11.84KK70 pKa = 10.02GGSRR74 pKa = 11.84KK75 pKa = 8.19QRR77 pKa = 11.84FTAGRR82 pKa = 11.84RR83 pKa = 11.84SARR86 pKa = 11.84QGVNRR91 pKa = 11.84LNRR94 pKa = 11.84RR95 pKa = 11.84KK96 pKa = 9.97NIQRR100 pKa = 11.84IAEE103 pKa = 4.08EE104 pKa = 3.88RR105 pKa = 11.84CTRR108 pKa = 11.84KK109 pKa = 8.44FTNLRR114 pKa = 11.84VLNSYY119 pKa = 8.44WVGEE123 pKa = 4.4DD124 pKa = 4.78GSQKK128 pKa = 8.97WFEE131 pKa = 4.62VIMLDD136 pKa = 3.77PEE138 pKa = 4.41HH139 pKa = 6.93PAIEE143 pKa = 4.75NDD145 pKa = 5.08DD146 pKa = 4.12DD147 pKa = 5.67LNWICDD153 pKa = 3.7DD154 pKa = 3.61THH156 pKa = 7.42KK157 pKa = 10.93NRR159 pKa = 11.84AMRR162 pKa = 11.84GLTSAGTQNRR172 pKa = 11.84GLEE175 pKa = 4.37SKK177 pKa = 11.04GKK179 pKa = 8.02GTEE182 pKa = 3.7RR183 pKa = 11.84TRR185 pKa = 11.84PSLSSNKK192 pKa = 9.22SRR194 pKa = 11.84KK195 pKa = 8.98
Molecular weight: 22.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
979672 |
24 |
1886 |
292.1 |
31.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.136 ± 0.055 | 0.768 ± 0.016 |
8.247 ± 0.053 | 8.918 ± 0.056 |
3.318 ± 0.028 | 8.128 ± 0.041 |
2.021 ± 0.024 | 4.267 ± 0.033 |
1.954 ± 0.028 | 8.963 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.779 ± 0.017 | 2.465 ± 0.023 |
4.452 ± 0.027 | 3.097 ± 0.024 |
6.248 ± 0.043 | 5.671 ± 0.037 |
6.9 ± 0.037 | 8.846 ± 0.047 |
1.107 ± 0.015 | 2.714 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
