
Torque teno virus 5
Taxonomy: Viruses; Anelloviridae; Alphatorquevirus
Average proteome isoelectric point is 7.58
Get precalculated fractions of proteins
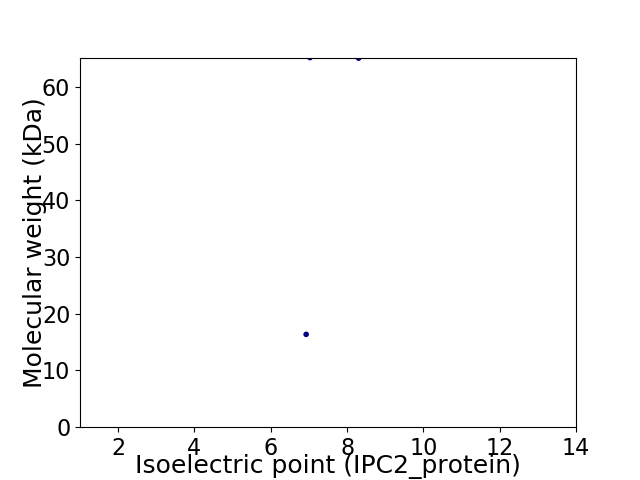
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q99AR2|Q99AR2_9VIRU Orf2 OS=Torque teno virus 5 OX=687344 PE=4 SV=1
MM1 pKa = 7.51GKK3 pKa = 9.73ALKK6 pKa = 10.2KK7 pKa = 9.37ATPLTMFFGRR17 pKa = 11.84VYY19 pKa = 10.22RR20 pKa = 11.84HH21 pKa = 5.62QKK23 pKa = 10.3RR24 pKa = 11.84KK25 pKa = 9.49VLLSLVHH32 pKa = 6.47TPQKK36 pKa = 9.78TPSSMNRR43 pKa = 11.84WSPPAHH49 pKa = 7.14DD50 pKa = 3.44ARR52 pKa = 11.84GIEE55 pKa = 4.17RR56 pKa = 11.84QWYY59 pKa = 7.26EE60 pKa = 4.34CVWRR64 pKa = 11.84SHH66 pKa = 6.67AACCGCGDD74 pKa = 4.16FVGHH78 pKa = 6.76INDD81 pKa = 3.67LAHH84 pKa = 7.44RR85 pKa = 11.84FGRR88 pKa = 11.84PPGSRR93 pKa = 11.84PPTTPQPPAVRR104 pKa = 11.84ALPALPAPEE113 pKa = 4.41PWSGAGGDD121 pKa = 3.91AGAGGDD127 pKa = 3.77GAAGRR132 pKa = 11.84GGGDD136 pKa = 3.23GDD138 pKa = 4.25AGDD141 pKa = 4.25VADD144 pKa = 5.55EE145 pKa = 4.81DD146 pKa = 4.86LLDD149 pKa = 4.37AVDD152 pKa = 3.91LAEE155 pKa = 4.18
MM1 pKa = 7.51GKK3 pKa = 9.73ALKK6 pKa = 10.2KK7 pKa = 9.37ATPLTMFFGRR17 pKa = 11.84VYY19 pKa = 10.22RR20 pKa = 11.84HH21 pKa = 5.62QKK23 pKa = 10.3RR24 pKa = 11.84KK25 pKa = 9.49VLLSLVHH32 pKa = 6.47TPQKK36 pKa = 9.78TPSSMNRR43 pKa = 11.84WSPPAHH49 pKa = 7.14DD50 pKa = 3.44ARR52 pKa = 11.84GIEE55 pKa = 4.17RR56 pKa = 11.84QWYY59 pKa = 7.26EE60 pKa = 4.34CVWRR64 pKa = 11.84SHH66 pKa = 6.67AACCGCGDD74 pKa = 4.16FVGHH78 pKa = 6.76INDD81 pKa = 3.67LAHH84 pKa = 7.44RR85 pKa = 11.84FGRR88 pKa = 11.84PPGSRR93 pKa = 11.84PPTTPQPPAVRR104 pKa = 11.84ALPALPAPEE113 pKa = 4.41PWSGAGGDD121 pKa = 3.91AGAGGDD127 pKa = 3.77GAAGRR132 pKa = 11.84GGGDD136 pKa = 3.23GDD138 pKa = 4.25AGDD141 pKa = 4.25VADD144 pKa = 5.55EE145 pKa = 4.81DD146 pKa = 4.86LLDD149 pKa = 4.37AVDD152 pKa = 3.91LAEE155 pKa = 4.18
Molecular weight: 16.37 kDa
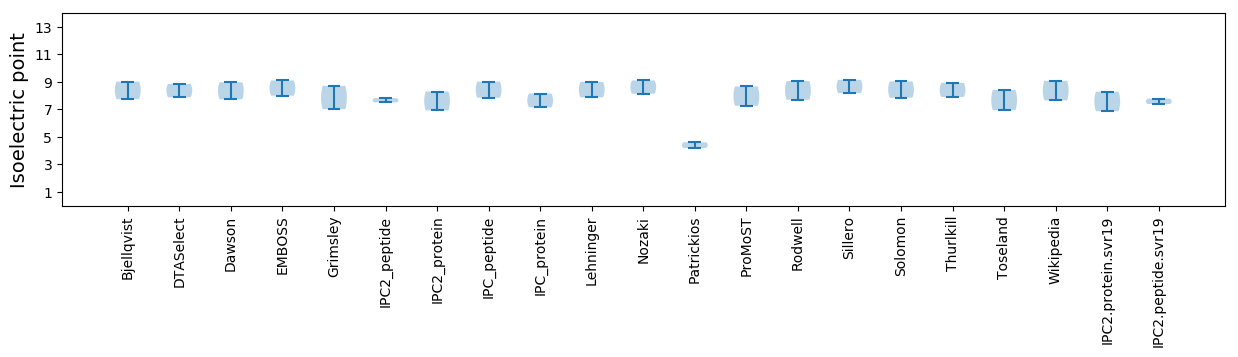
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q99AR2|Q99AR2_9VIRU Orf2 OS=Torque teno virus 5 OX=687344 PE=4 SV=1
MM1 pKa = 7.72LMLAKK6 pKa = 9.96KK7 pKa = 10.38RR8 pKa = 11.84IIIPSLKK15 pKa = 8.83TRR17 pKa = 11.84PSKK20 pKa = 10.44KK21 pKa = 9.58HH22 pKa = 5.16YY23 pKa = 10.16IKK25 pKa = 10.58IKK27 pKa = 10.09IGAPKK32 pKa = 10.3LFEE35 pKa = 5.04DD36 pKa = 3.05KK37 pKa = 10.15WYY39 pKa = 10.25SQRR42 pKa = 11.84DD43 pKa = 3.58LCDD46 pKa = 3.02TTLVVIEE53 pKa = 4.5ATVADD58 pKa = 4.09LQYY61 pKa = 11.06PFGSPLTNSICCNFQVLNSNYY82 pKa = 10.57DD83 pKa = 3.38NALSTLIGTQGDD95 pKa = 3.74KK96 pKa = 11.28DD97 pKa = 3.51RR98 pKa = 11.84DD99 pKa = 3.41QCYY102 pKa = 10.3KK103 pKa = 10.75FLLNTITYY111 pKa = 9.08YY112 pKa = 9.67NTSQTLAQLNKK123 pKa = 9.92FLPTTNTTQLSQNNAQNSINPSNTTEE149 pKa = 3.74NTYY152 pKa = 11.12DD153 pKa = 3.35SRR155 pKa = 11.84NTEE158 pKa = 3.41IHH160 pKa = 6.6NNTLYY165 pKa = 11.09GGAAYY170 pKa = 10.14YY171 pKa = 9.33KK172 pKa = 10.57QNGSNTNTLPPHH184 pKa = 6.66KK185 pKa = 9.55MKK187 pKa = 10.48EE188 pKa = 3.98ANKK191 pKa = 9.59TYY193 pKa = 9.58TAAAKK198 pKa = 10.24KK199 pKa = 10.15ILSNWNFTNMSTSSSADD216 pKa = 3.52LQGLNYY222 pKa = 9.68FDD224 pKa = 5.71YY225 pKa = 8.65YY226 pKa = 11.15TGMFSSIFLASGRR239 pKa = 11.84SNWEE243 pKa = 3.63VKK245 pKa = 10.52GSYY248 pKa = 9.75TDD250 pKa = 2.89ITYY253 pKa = 11.05NPLVDD258 pKa = 3.66KK259 pKa = 11.61GEE261 pKa = 4.46GNMIWIDD268 pKa = 4.22WITKK272 pKa = 9.87GDD274 pKa = 3.87TIYY277 pKa = 11.05SEE279 pKa = 4.67NKK281 pKa = 9.78SKK283 pKa = 10.94CLLRR287 pKa = 11.84DD288 pKa = 3.29LPMWALCYY296 pKa = 10.44GYY298 pKa = 10.99ADD300 pKa = 3.87YY301 pKa = 11.32VLKK304 pKa = 9.93CTGISSIKK312 pKa = 9.53NEE314 pKa = 3.42ARR316 pKa = 11.84IVMRR320 pKa = 11.84CPYY323 pKa = 9.66TYY325 pKa = 10.17PQLVKK330 pKa = 10.8HH331 pKa = 6.27NNDD334 pKa = 2.62NFGFVVYY341 pKa = 10.42SDD343 pKa = 3.17NFGRR347 pKa = 11.84GRR349 pKa = 11.84MPGGDD354 pKa = 3.64PVPSTRR360 pKa = 11.84NRR362 pKa = 11.84VHH364 pKa = 6.63WYY366 pKa = 8.13ITITHH371 pKa = 4.63QTEE374 pKa = 3.95VLEE377 pKa = 5.43CISQTGPFAYY387 pKa = 10.27HH388 pKa = 5.62SDD390 pKa = 3.19EE391 pKa = 4.26RR392 pKa = 11.84KK393 pKa = 9.74AVLTIKK399 pKa = 11.01YY400 pKa = 9.18NFRR403 pKa = 11.84WKK405 pKa = 9.9WGGNPVFQQILRR417 pKa = 11.84DD418 pKa = 3.9PCSGSPGSGPRR429 pKa = 11.84RR430 pKa = 11.84VPRR433 pKa = 11.84SIQVDD438 pKa = 3.58DD439 pKa = 4.86PKK441 pKa = 11.22YY442 pKa = 9.11QTPEE446 pKa = 4.46YY447 pKa = 9.44IWHH450 pKa = 6.44AWDD453 pKa = 3.81FRR455 pKa = 11.84RR456 pKa = 11.84GLFSQKK462 pKa = 10.07GIKK465 pKa = 9.54RR466 pKa = 11.84VSEE469 pKa = 3.96QPTDD473 pKa = 3.08VDD475 pKa = 4.27LPTGRR480 pKa = 11.84GKK482 pKa = 10.52RR483 pKa = 11.84PKK485 pKa = 9.69RR486 pKa = 11.84DD487 pKa = 2.94TGGPQEE493 pKa = 4.14QGQEE497 pKa = 4.18EE498 pKa = 5.05GSSSIRR504 pKa = 11.84RR505 pKa = 11.84VLQQWLHH512 pKa = 5.8SSQEE516 pKa = 3.85QSQEE520 pKa = 4.19SEE522 pKa = 4.17EE523 pKa = 4.17EE524 pKa = 3.94AAPTTLSQEE533 pKa = 4.03LQKK536 pKa = 10.58QLKK539 pKa = 8.04QQQLMGKK546 pKa = 7.26QLRR549 pKa = 11.84EE550 pKa = 4.1LSLQLAQVQAGGHH563 pKa = 5.81LHH565 pKa = 7.38PLLQCHH571 pKa = 6.21AA572 pKa = 4.67
MM1 pKa = 7.72LMLAKK6 pKa = 9.96KK7 pKa = 10.38RR8 pKa = 11.84IIIPSLKK15 pKa = 8.83TRR17 pKa = 11.84PSKK20 pKa = 10.44KK21 pKa = 9.58HH22 pKa = 5.16YY23 pKa = 10.16IKK25 pKa = 10.58IKK27 pKa = 10.09IGAPKK32 pKa = 10.3LFEE35 pKa = 5.04DD36 pKa = 3.05KK37 pKa = 10.15WYY39 pKa = 10.25SQRR42 pKa = 11.84DD43 pKa = 3.58LCDD46 pKa = 3.02TTLVVIEE53 pKa = 4.5ATVADD58 pKa = 4.09LQYY61 pKa = 11.06PFGSPLTNSICCNFQVLNSNYY82 pKa = 10.57DD83 pKa = 3.38NALSTLIGTQGDD95 pKa = 3.74KK96 pKa = 11.28DD97 pKa = 3.51RR98 pKa = 11.84DD99 pKa = 3.41QCYY102 pKa = 10.3KK103 pKa = 10.75FLLNTITYY111 pKa = 9.08YY112 pKa = 9.67NTSQTLAQLNKK123 pKa = 9.92FLPTTNTTQLSQNNAQNSINPSNTTEE149 pKa = 3.74NTYY152 pKa = 11.12DD153 pKa = 3.35SRR155 pKa = 11.84NTEE158 pKa = 3.41IHH160 pKa = 6.6NNTLYY165 pKa = 11.09GGAAYY170 pKa = 10.14YY171 pKa = 9.33KK172 pKa = 10.57QNGSNTNTLPPHH184 pKa = 6.66KK185 pKa = 9.55MKK187 pKa = 10.48EE188 pKa = 3.98ANKK191 pKa = 9.59TYY193 pKa = 9.58TAAAKK198 pKa = 10.24KK199 pKa = 10.15ILSNWNFTNMSTSSSADD216 pKa = 3.52LQGLNYY222 pKa = 9.68FDD224 pKa = 5.71YY225 pKa = 8.65YY226 pKa = 11.15TGMFSSIFLASGRR239 pKa = 11.84SNWEE243 pKa = 3.63VKK245 pKa = 10.52GSYY248 pKa = 9.75TDD250 pKa = 2.89ITYY253 pKa = 11.05NPLVDD258 pKa = 3.66KK259 pKa = 11.61GEE261 pKa = 4.46GNMIWIDD268 pKa = 4.22WITKK272 pKa = 9.87GDD274 pKa = 3.87TIYY277 pKa = 11.05SEE279 pKa = 4.67NKK281 pKa = 9.78SKK283 pKa = 10.94CLLRR287 pKa = 11.84DD288 pKa = 3.29LPMWALCYY296 pKa = 10.44GYY298 pKa = 10.99ADD300 pKa = 3.87YY301 pKa = 11.32VLKK304 pKa = 9.93CTGISSIKK312 pKa = 9.53NEE314 pKa = 3.42ARR316 pKa = 11.84IVMRR320 pKa = 11.84CPYY323 pKa = 9.66TYY325 pKa = 10.17PQLVKK330 pKa = 10.8HH331 pKa = 6.27NNDD334 pKa = 2.62NFGFVVYY341 pKa = 10.42SDD343 pKa = 3.17NFGRR347 pKa = 11.84GRR349 pKa = 11.84MPGGDD354 pKa = 3.64PVPSTRR360 pKa = 11.84NRR362 pKa = 11.84VHH364 pKa = 6.63WYY366 pKa = 8.13ITITHH371 pKa = 4.63QTEE374 pKa = 3.95VLEE377 pKa = 5.43CISQTGPFAYY387 pKa = 10.27HH388 pKa = 5.62SDD390 pKa = 3.19EE391 pKa = 4.26RR392 pKa = 11.84KK393 pKa = 9.74AVLTIKK399 pKa = 11.01YY400 pKa = 9.18NFRR403 pKa = 11.84WKK405 pKa = 9.9WGGNPVFQQILRR417 pKa = 11.84DD418 pKa = 3.9PCSGSPGSGPRR429 pKa = 11.84RR430 pKa = 11.84VPRR433 pKa = 11.84SIQVDD438 pKa = 3.58DD439 pKa = 4.86PKK441 pKa = 11.22YY442 pKa = 9.11QTPEE446 pKa = 4.46YY447 pKa = 9.44IWHH450 pKa = 6.44AWDD453 pKa = 3.81FRR455 pKa = 11.84RR456 pKa = 11.84GLFSQKK462 pKa = 10.07GIKK465 pKa = 9.54RR466 pKa = 11.84VSEE469 pKa = 3.96QPTDD473 pKa = 3.08VDD475 pKa = 4.27LPTGRR480 pKa = 11.84GKK482 pKa = 10.52RR483 pKa = 11.84PKK485 pKa = 9.69RR486 pKa = 11.84DD487 pKa = 2.94TGGPQEE493 pKa = 4.14QGQEE497 pKa = 4.18EE498 pKa = 5.05GSSSIRR504 pKa = 11.84RR505 pKa = 11.84VLQQWLHH512 pKa = 5.8SSQEE516 pKa = 3.85QSQEE520 pKa = 4.19SEE522 pKa = 4.17EE523 pKa = 4.17EE524 pKa = 3.94AAPTTLSQEE533 pKa = 4.03LQKK536 pKa = 10.58QLKK539 pKa = 8.04QQQLMGKK546 pKa = 7.26QLRR549 pKa = 11.84EE550 pKa = 4.1LSLQLAQVQAGGHH563 pKa = 5.81LHH565 pKa = 7.38PLLQCHH571 pKa = 6.21AA572 pKa = 4.67
Molecular weight: 65.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
727 |
155 |
572 |
363.5 |
40.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.327 ± 3.153 | 2.063 ± 0.248 |
5.502 ± 1.074 | 3.989 ± 0.366 |
2.889 ± 0.148 | 8.116 ± 2.605 |
2.476 ± 0.669 | 4.539 ± 1.558 |
5.915 ± 0.98 | 8.116 ± 0.488 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.788 ± 0.071 | 5.502 ± 2.019 |
6.327 ± 1.916 | 6.052 ± 1.665 |
5.502 ± 1.074 | 7.29 ± 1.33 |
7.015 ± 1.508 | 4.127 ± 0.496 |
2.201 ± 0.182 | 4.264 ± 1.426 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
