
Organic Lake virophage
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Maveriviricetes; Priklausovirales; Lavidaviridae; unclassified Lavidaviridae
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 24 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
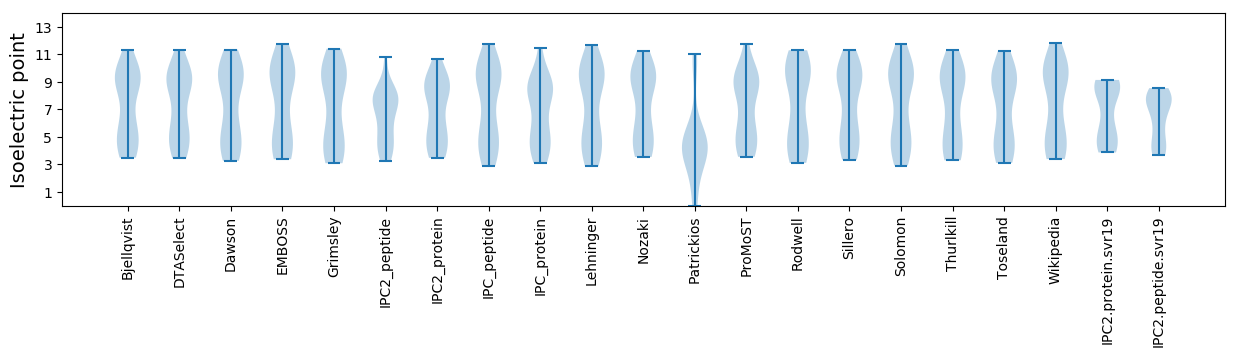
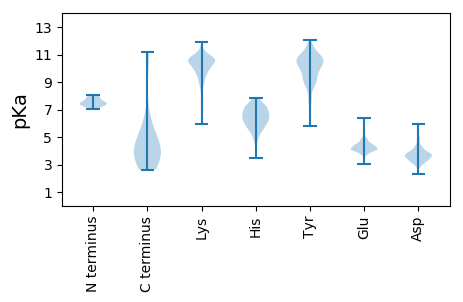
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|F2Y0M5|F2Y0M5_9VIRU Uncharacterized protein OS=Organic Lake virophage OX=938080 GN=OLV21 PE=4 SV=1
MM1 pKa = 7.55FNLTDD6 pKa = 4.01DD7 pKa = 4.16SKK9 pKa = 11.15VTILKK14 pKa = 9.77SGNYY18 pKa = 8.81KK19 pKa = 10.18VEE21 pKa = 4.33FMCGFFNVSKK31 pKa = 10.51GRR33 pKa = 11.84ANIRR37 pKa = 11.84VGCRR41 pKa = 11.84INGIYY46 pKa = 10.52DD47 pKa = 3.41KK48 pKa = 10.85TFGGQPSCYY57 pKa = 9.88LRR59 pKa = 11.84DD60 pKa = 3.5QAYY63 pKa = 9.78VRR65 pKa = 11.84YY66 pKa = 9.13GSCSNSLYY74 pKa = 10.73FSLNANDD81 pKa = 4.34TIEE84 pKa = 5.06LEE86 pKa = 4.49SNLNLGDD93 pKa = 4.21EE94 pKa = 4.65IGFDD98 pKa = 4.2SNFDD102 pKa = 3.58SAFQLLRR109 pKa = 11.84GSNILITYY117 pKa = 9.56LDD119 pKa = 3.44QTGPQGDD126 pKa = 3.9TGATGADD133 pKa = 4.25GIDD136 pKa = 4.56GIDD139 pKa = 3.65GATGATGPQGIQGEE153 pKa = 4.54TGADD157 pKa = 3.53GADD160 pKa = 3.73GADD163 pKa = 3.85GADD166 pKa = 3.9GATGPTGPTGVDD178 pKa = 3.29GADD181 pKa = 3.66GATGPQGIQGEE192 pKa = 4.49TGPQGIQGEE201 pKa = 4.54TGADD205 pKa = 3.59GADD208 pKa = 3.71GAIGPTGPAGADD220 pKa = 3.56GEE222 pKa = 4.69VTLAQLNTKK231 pKa = 10.11QDD233 pKa = 3.29VLTAGNNITIDD244 pKa = 3.7VNNVISSSGGEE255 pKa = 4.01GGGGITQQEE264 pKa = 4.4LDD266 pKa = 4.66DD267 pKa = 4.44GLEE270 pKa = 4.11TKK272 pKa = 10.36QDD274 pKa = 3.3ILTAGANISIVGSTINSGSSAYY296 pKa = 9.76FLCFLNLNYY305 pKa = 10.39TNLILGDD312 pKa = 3.72YY313 pKa = 10.65ARR315 pKa = 11.84FPAISFQKK323 pKa = 10.16PNTGTMVEE331 pKa = 3.88QHH333 pKa = 6.65RR334 pKa = 11.84GHH336 pKa = 7.48AYY338 pKa = 8.99TIQEE342 pKa = 4.06TGLYY346 pKa = 9.92LIGYY350 pKa = 8.15SLTMLNQGGTTAIQIVYY367 pKa = 9.23VRR369 pKa = 11.84NGVEE373 pKa = 4.89KK374 pKa = 10.65SITYY378 pKa = 10.38NGITIPTSEE387 pKa = 4.16NRR389 pKa = 11.84SIIFPLEE396 pKa = 4.21AGDD399 pKa = 5.14LIAVKK404 pKa = 10.25YY405 pKa = 10.49KK406 pKa = 10.59SGNAGYY412 pKa = 8.1LTLYY416 pKa = 7.5GTPSTEE422 pKa = 4.06NIQTNMYY429 pKa = 9.68GYY431 pKa = 10.62RR432 pKa = 11.84IAA434 pKa = 5.59
MM1 pKa = 7.55FNLTDD6 pKa = 4.01DD7 pKa = 4.16SKK9 pKa = 11.15VTILKK14 pKa = 9.77SGNYY18 pKa = 8.81KK19 pKa = 10.18VEE21 pKa = 4.33FMCGFFNVSKK31 pKa = 10.51GRR33 pKa = 11.84ANIRR37 pKa = 11.84VGCRR41 pKa = 11.84INGIYY46 pKa = 10.52DD47 pKa = 3.41KK48 pKa = 10.85TFGGQPSCYY57 pKa = 9.88LRR59 pKa = 11.84DD60 pKa = 3.5QAYY63 pKa = 9.78VRR65 pKa = 11.84YY66 pKa = 9.13GSCSNSLYY74 pKa = 10.73FSLNANDD81 pKa = 4.34TIEE84 pKa = 5.06LEE86 pKa = 4.49SNLNLGDD93 pKa = 4.21EE94 pKa = 4.65IGFDD98 pKa = 4.2SNFDD102 pKa = 3.58SAFQLLRR109 pKa = 11.84GSNILITYY117 pKa = 9.56LDD119 pKa = 3.44QTGPQGDD126 pKa = 3.9TGATGADD133 pKa = 4.25GIDD136 pKa = 4.56GIDD139 pKa = 3.65GATGATGPQGIQGEE153 pKa = 4.54TGADD157 pKa = 3.53GADD160 pKa = 3.73GADD163 pKa = 3.85GADD166 pKa = 3.9GATGPTGPTGVDD178 pKa = 3.29GADD181 pKa = 3.66GATGPQGIQGEE192 pKa = 4.49TGPQGIQGEE201 pKa = 4.54TGADD205 pKa = 3.59GADD208 pKa = 3.71GAIGPTGPAGADD220 pKa = 3.56GEE222 pKa = 4.69VTLAQLNTKK231 pKa = 10.11QDD233 pKa = 3.29VLTAGNNITIDD244 pKa = 3.7VNNVISSSGGEE255 pKa = 4.01GGGGITQQEE264 pKa = 4.4LDD266 pKa = 4.66DD267 pKa = 4.44GLEE270 pKa = 4.11TKK272 pKa = 10.36QDD274 pKa = 3.3ILTAGANISIVGSTINSGSSAYY296 pKa = 9.76FLCFLNLNYY305 pKa = 10.39TNLILGDD312 pKa = 3.72YY313 pKa = 10.65ARR315 pKa = 11.84FPAISFQKK323 pKa = 10.16PNTGTMVEE331 pKa = 3.88QHH333 pKa = 6.65RR334 pKa = 11.84GHH336 pKa = 7.48AYY338 pKa = 8.99TIQEE342 pKa = 4.06TGLYY346 pKa = 9.92LIGYY350 pKa = 8.15SLTMLNQGGTTAIQIVYY367 pKa = 9.23VRR369 pKa = 11.84NGVEE373 pKa = 4.89KK374 pKa = 10.65SITYY378 pKa = 10.38NGITIPTSEE387 pKa = 4.16NRR389 pKa = 11.84SIIFPLEE396 pKa = 4.21AGDD399 pKa = 5.14LIAVKK404 pKa = 10.25YY405 pKa = 10.49KK406 pKa = 10.59SGNAGYY412 pKa = 8.1LTLYY416 pKa = 7.5GTPSTEE422 pKa = 4.06NIQTNMYY429 pKa = 9.68GYY431 pKa = 10.62RR432 pKa = 11.84IAA434 pKa = 5.59
Molecular weight: 45.05 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F2Y0M1|F2Y0M1_9VIRU Uncharacterized protein OS=Organic Lake virophage OX=938080 GN=OLV16 PE=4 SV=1
MM1 pKa = 7.3RR2 pKa = 11.84TSSTLRR8 pKa = 11.84TSSTSSTNSTISPINSISPSRR29 pKa = 11.84TLRR32 pKa = 11.84TSSTSLSLRR41 pKa = 11.84TLWTSSTINAISTSSTSSTINTINTSRR68 pKa = 11.84TSFALYY74 pKa = 9.08TLWTSSTINTINTSRR89 pKa = 11.84TSFALYY95 pKa = 9.08TLWTSSTINTISTLWTSSTSSTSSTINTVNSISTSFALYY134 pKa = 9.1TLRR137 pKa = 11.84TSFALYY143 pKa = 9.08TLWTSSTSSTSSTINTVNSISTSFALYY170 pKa = 9.1TLRR173 pKa = 11.84TSFALYY179 pKa = 9.08TLWTSSTSSTSSTSSTSSTNSTVSTVNSISTSSTSRR215 pKa = 11.84TLRR218 pKa = 11.84TSSTSSTNSTSSTNSTVNSISSISTSRR245 pKa = 11.84TSSTISSISSISSISSSRR263 pKa = 11.84TSRR266 pKa = 11.84TSFALYY272 pKa = 9.08TLWTSSTINPISTISTISTSFALYY296 pKa = 9.1TLRR299 pKa = 11.84TSFALYY305 pKa = 9.13TLRR308 pKa = 11.84TSSTISPISPISTISPISTISTSRR332 pKa = 11.84TSRR335 pKa = 11.84TSFALYY341 pKa = 9.08TLWTSFSYY349 pKa = 11.3VPFFSCRR356 pKa = 11.84RR357 pKa = 11.84NYY359 pKa = 9.77ISCLINRR366 pKa = 11.84YY367 pKa = 9.46LCTGYY372 pKa = 11.21
MM1 pKa = 7.3RR2 pKa = 11.84TSSTLRR8 pKa = 11.84TSSTSSTNSTISPINSISPSRR29 pKa = 11.84TLRR32 pKa = 11.84TSSTSLSLRR41 pKa = 11.84TLWTSSTINAISTSSTSSTINTINTSRR68 pKa = 11.84TSFALYY74 pKa = 9.08TLWTSSTINTINTSRR89 pKa = 11.84TSFALYY95 pKa = 9.08TLWTSSTINTISTLWTSSTSSTSSTINTVNSISTSFALYY134 pKa = 9.1TLRR137 pKa = 11.84TSFALYY143 pKa = 9.08TLWTSSTSSTSSTINTVNSISTSFALYY170 pKa = 9.1TLRR173 pKa = 11.84TSFALYY179 pKa = 9.08TLWTSSTSSTSSTSSTSSTNSTVSTVNSISTSSTSRR215 pKa = 11.84TLRR218 pKa = 11.84TSSTSSTNSTSSTNSTVNSISSISTSRR245 pKa = 11.84TSSTISSISSISSISSSRR263 pKa = 11.84TSRR266 pKa = 11.84TSFALYY272 pKa = 9.08TLWTSSTINPISTISTISTSFALYY296 pKa = 9.1TLRR299 pKa = 11.84TSFALYY305 pKa = 9.13TLRR308 pKa = 11.84TSSTISPISPISTISPISTISTSRR332 pKa = 11.84TSRR335 pKa = 11.84TSFALYY341 pKa = 9.08TLWTSFSYY349 pKa = 11.3VPFFSCRR356 pKa = 11.84RR357 pKa = 11.84NYY359 pKa = 9.77ISCLINRR366 pKa = 11.84YY367 pKa = 9.46LCTGYY372 pKa = 11.21
Molecular weight: 39.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8420 |
96 |
1036 |
350.8 |
38.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.903 ± 0.671 | 1.247 ± 0.303 |
5.95 ± 0.545 | 5.523 ± 0.507 |
3.349 ± 0.382 | 7.518 ± 1.449 |
1.366 ± 0.224 | 7.031 ± 0.359 |
6.722 ± 1.095 | 8.29 ± 0.672 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.067 ± 0.276 | 6.354 ± 0.465 |
4.537 ± 1.028 | 3.99 ± 0.497 |
2.922 ± 0.258 | 9.204 ± 1.144 |
8.029 ± 0.805 | 5.475 ± 0.562 |
0.665 ± 0.135 | 3.86 ± 0.436 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
