
Capybara microvirus Cap1_SP_99
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.76
Get precalculated fractions of proteins
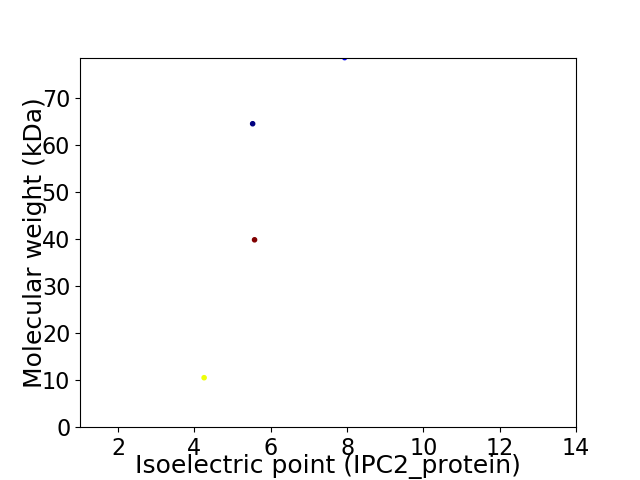
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W7P6|A0A4P8W7P6_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_99 OX=2584802 PE=4 SV=1
MM1 pKa = 7.79ILRR4 pKa = 11.84KK5 pKa = 9.69GVNDD9 pKa = 3.61TQTRR13 pKa = 11.84PIGTTVYY20 pKa = 10.06PPTLIVRR27 pKa = 11.84TYY29 pKa = 9.83VNEE32 pKa = 3.86EE33 pKa = 4.15GEE35 pKa = 4.3EE36 pKa = 4.36VSCVEE41 pKa = 4.55SLDD44 pKa = 5.1DD45 pKa = 5.86IPFEE49 pKa = 4.35DD50 pKa = 5.06ANVGDD55 pKa = 4.16YY56 pKa = 10.71TLDD59 pKa = 3.53ALFKK63 pKa = 10.97AGINPRR69 pKa = 11.84SVNFNSTVSNVDD81 pKa = 2.93LHH83 pKa = 7.28DD84 pKa = 4.62ALNEE88 pKa = 3.89INNLKK93 pKa = 10.35VEE95 pKa = 4.2
MM1 pKa = 7.79ILRR4 pKa = 11.84KK5 pKa = 9.69GVNDD9 pKa = 3.61TQTRR13 pKa = 11.84PIGTTVYY20 pKa = 10.06PPTLIVRR27 pKa = 11.84TYY29 pKa = 9.83VNEE32 pKa = 3.86EE33 pKa = 4.15GEE35 pKa = 4.3EE36 pKa = 4.36VSCVEE41 pKa = 4.55SLDD44 pKa = 5.1DD45 pKa = 5.86IPFEE49 pKa = 4.35DD50 pKa = 5.06ANVGDD55 pKa = 4.16YY56 pKa = 10.71TLDD59 pKa = 3.53ALFKK63 pKa = 10.97AGINPRR69 pKa = 11.84SVNFNSTVSNVDD81 pKa = 2.93LHH83 pKa = 7.28DD84 pKa = 4.62ALNEE88 pKa = 3.89INNLKK93 pKa = 10.35VEE95 pKa = 4.2
Molecular weight: 10.53 kDa
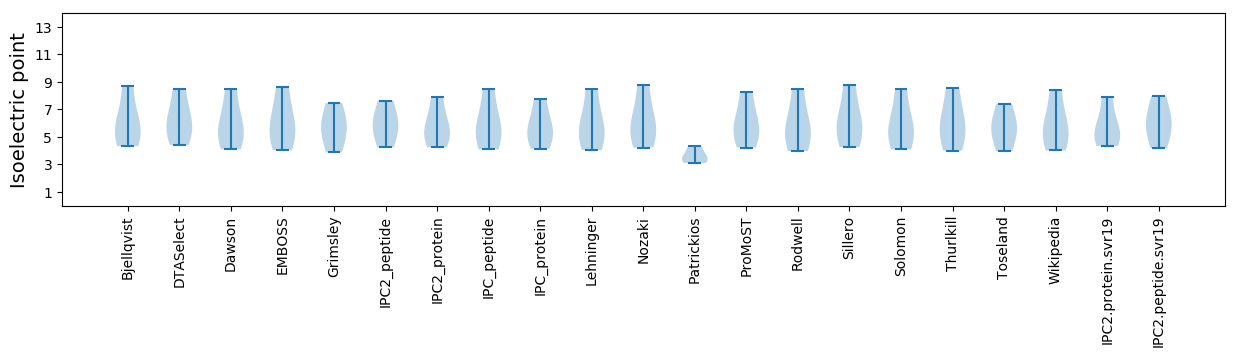
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W4D4|A0A4P8W4D4_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_99 OX=2584802 PE=3 SV=1
MM1 pKa = 7.71ISDD4 pKa = 4.66SYY6 pKa = 10.71ICSSIPLRR14 pKa = 11.84SYY16 pKa = 11.35YY17 pKa = 10.38YY18 pKa = 10.67DD19 pKa = 3.49CFRR22 pKa = 11.84DD23 pKa = 3.56KK24 pKa = 11.18SIIRR28 pKa = 11.84VARR31 pKa = 11.84RR32 pKa = 11.84EE33 pKa = 3.97NFGTSQLAQKK43 pKa = 10.3EE44 pKa = 4.37YY45 pKa = 10.01TLRR48 pKa = 11.84LYY50 pKa = 11.04SEE52 pKa = 4.95LKK54 pKa = 8.98RR55 pKa = 11.84TYY57 pKa = 9.76ALHH60 pKa = 5.53GRR62 pKa = 11.84VYY64 pKa = 10.43FYY66 pKa = 10.62TLTYY70 pKa = 10.94SDD72 pKa = 4.09VNLPKK77 pKa = 10.68YY78 pKa = 10.42KK79 pKa = 10.48GVSCVDD85 pKa = 3.55YY86 pKa = 10.71NHH88 pKa = 6.42IRR90 pKa = 11.84AMFHH94 pKa = 6.87DD95 pKa = 5.29FLYY98 pKa = 9.39EE99 pKa = 3.74QCEE102 pKa = 3.97RR103 pKa = 11.84AHH105 pKa = 6.2CAKK108 pKa = 10.07IKK110 pKa = 10.02YY111 pKa = 9.52FVVTEE116 pKa = 3.98YY117 pKa = 11.6GDD119 pKa = 3.84GKK121 pKa = 10.68GSRR124 pKa = 11.84GKK126 pKa = 10.72GNNPHH131 pKa = 5.87FHH133 pKa = 6.73VIFMFQPHH141 pKa = 6.57ANVGKK146 pKa = 9.82HH147 pKa = 3.7LTGRR151 pKa = 11.84KK152 pKa = 8.77KK153 pKa = 10.62SYY155 pKa = 10.27MNIFFPPTPHH165 pKa = 7.1QMLQYY170 pKa = 9.7MRR172 pKa = 11.84YY173 pKa = 7.91YY174 pKa = 8.84WQGNKK179 pKa = 7.5ITNVYY184 pKa = 10.08HH185 pKa = 7.4DD186 pKa = 3.95GSFDD190 pKa = 3.53VEE192 pKa = 4.96SYY194 pKa = 11.0KK195 pKa = 10.41RR196 pKa = 11.84WQDD199 pKa = 3.19LKK201 pKa = 11.32YY202 pKa = 10.78GFVKK206 pKa = 10.46PGCLKK211 pKa = 10.36GARR214 pKa = 11.84NGAACGEE221 pKa = 4.15VLPDD225 pKa = 4.27NIDD228 pKa = 3.28ALGYY232 pKa = 6.09VTKK235 pKa = 10.88YY236 pKa = 6.8VTKK239 pKa = 8.83DD240 pKa = 2.86THH242 pKa = 5.36KK243 pKa = 10.63RR244 pKa = 11.84RR245 pKa = 11.84VEE247 pKa = 3.75TEE249 pKa = 3.55IYY251 pKa = 9.59RR252 pKa = 11.84RR253 pKa = 11.84IYY255 pKa = 9.63GEE257 pKa = 4.65SYY259 pKa = 10.58FDD261 pKa = 3.94CANNLDD267 pKa = 3.75LVYY270 pKa = 10.44RR271 pKa = 11.84CLRR274 pKa = 11.84FSVEE278 pKa = 4.07EE279 pKa = 4.01KK280 pKa = 10.44NKK282 pKa = 10.05EE283 pKa = 3.84FASEE287 pKa = 4.27YY288 pKa = 10.58EE289 pKa = 4.33DD290 pKa = 3.38IKK292 pKa = 11.57GDD294 pKa = 3.58VNYY297 pKa = 10.77DD298 pKa = 3.78GLSSIYY304 pKa = 10.81DD305 pKa = 3.52NLADD309 pKa = 3.65VGKK312 pKa = 10.95LEE314 pKa = 4.43FSDD317 pKa = 4.93SITEE321 pKa = 4.28KK322 pKa = 10.88LCSAYY327 pKa = 10.59DD328 pKa = 3.79NYY330 pKa = 11.35LHH332 pKa = 6.59SCSSRR337 pKa = 11.84NVSPYY342 pKa = 10.34FFTYY346 pKa = 10.56CMNKK350 pKa = 9.46GVRR353 pKa = 11.84LLFPSLRR360 pKa = 11.84DD361 pKa = 3.5VQLNRR366 pKa = 11.84IKK368 pKa = 10.91HH369 pKa = 5.61LFNKK373 pKa = 8.96VFHH376 pKa = 6.9SMLEE380 pKa = 4.2EE381 pKa = 4.01YY382 pKa = 10.71QNEE385 pKa = 4.23CVKK388 pKa = 10.75YY389 pKa = 9.21YY390 pKa = 10.61RR391 pKa = 11.84NRR393 pKa = 11.84YY394 pKa = 5.1STKK397 pKa = 9.91VRR399 pKa = 11.84ISQHH403 pKa = 5.73FGDD406 pKa = 5.21SIVEE410 pKa = 4.1DD411 pKa = 4.36FRR413 pKa = 11.84EE414 pKa = 3.82KK415 pKa = 10.81ALQNRR420 pKa = 11.84EE421 pKa = 3.97LVYY424 pKa = 11.02LLNINGKK431 pKa = 8.95VNQKK435 pKa = 9.48YY436 pKa = 8.83YY437 pKa = 10.5CGSYY441 pKa = 10.05YY442 pKa = 10.11YY443 pKa = 10.43RR444 pKa = 11.84KK445 pKa = 10.12LFYY448 pKa = 10.61KK449 pKa = 9.88VVKK452 pKa = 10.47GHH454 pKa = 7.06DD455 pKa = 3.56GSNKK459 pKa = 9.7YY460 pKa = 9.86VLNDD464 pKa = 3.44FGIEE468 pKa = 4.01YY469 pKa = 10.74KK470 pKa = 10.25MKK472 pKa = 10.0TLDD475 pKa = 4.61DD476 pKa = 4.52NIEE479 pKa = 4.03RR480 pKa = 11.84QFAKK484 pKa = 10.05YY485 pKa = 10.48KK486 pKa = 10.26RR487 pKa = 11.84MLVSVYY493 pKa = 10.54NDD495 pKa = 3.22CNLYY499 pKa = 9.28STFNDD504 pKa = 2.71WRR506 pKa = 11.84KK507 pKa = 10.52KK508 pKa = 9.69YY509 pKa = 10.47GYY511 pKa = 10.32DD512 pKa = 3.39YY513 pKa = 10.82GISFAGSFFSTLSSIGFSEE532 pKa = 3.88ITEE535 pKa = 4.45DD536 pKa = 4.81DD537 pKa = 4.12LLKK540 pKa = 10.81AAIFKK545 pKa = 10.07IVYY548 pKa = 8.75RR549 pKa = 11.84DD550 pKa = 3.48RR551 pKa = 11.84YY552 pKa = 7.43TDD554 pKa = 3.42CVGINHH560 pKa = 6.01VDD562 pKa = 3.13YY563 pKa = 8.24FTRR566 pKa = 11.84YY567 pKa = 7.76RR568 pKa = 11.84DD569 pKa = 3.62YY570 pKa = 10.65YY571 pKa = 8.6EE572 pKa = 5.3SYY574 pKa = 10.37IIPHH578 pKa = 6.04LVEE581 pKa = 4.85NYY583 pKa = 10.58SSLTSLYY590 pKa = 9.85DD591 pKa = 3.3INMKK595 pKa = 7.73TVCAYY600 pKa = 9.19PLSDD604 pKa = 3.5NVVLSPYY611 pKa = 10.95SKK613 pKa = 10.99LCDD616 pKa = 3.52LFEE619 pKa = 4.83ALNDD623 pKa = 3.8YY624 pKa = 10.33IVYY627 pKa = 10.34LRR629 pKa = 11.84DD630 pKa = 3.62IQWSNDD636 pKa = 3.09YY637 pKa = 10.57EE638 pKa = 4.23EE639 pKa = 4.86RR640 pKa = 11.84SRR642 pKa = 11.84LRR644 pKa = 11.84KK645 pKa = 8.48LHH647 pKa = 6.09KK648 pKa = 9.66RR649 pKa = 11.84SEE651 pKa = 4.13VEE653 pKa = 3.92EE654 pKa = 3.96YY655 pKa = 10.9LVANRR660 pKa = 11.84YY661 pKa = 7.76LL662 pKa = 3.59
MM1 pKa = 7.71ISDD4 pKa = 4.66SYY6 pKa = 10.71ICSSIPLRR14 pKa = 11.84SYY16 pKa = 11.35YY17 pKa = 10.38YY18 pKa = 10.67DD19 pKa = 3.49CFRR22 pKa = 11.84DD23 pKa = 3.56KK24 pKa = 11.18SIIRR28 pKa = 11.84VARR31 pKa = 11.84RR32 pKa = 11.84EE33 pKa = 3.97NFGTSQLAQKK43 pKa = 10.3EE44 pKa = 4.37YY45 pKa = 10.01TLRR48 pKa = 11.84LYY50 pKa = 11.04SEE52 pKa = 4.95LKK54 pKa = 8.98RR55 pKa = 11.84TYY57 pKa = 9.76ALHH60 pKa = 5.53GRR62 pKa = 11.84VYY64 pKa = 10.43FYY66 pKa = 10.62TLTYY70 pKa = 10.94SDD72 pKa = 4.09VNLPKK77 pKa = 10.68YY78 pKa = 10.42KK79 pKa = 10.48GVSCVDD85 pKa = 3.55YY86 pKa = 10.71NHH88 pKa = 6.42IRR90 pKa = 11.84AMFHH94 pKa = 6.87DD95 pKa = 5.29FLYY98 pKa = 9.39EE99 pKa = 3.74QCEE102 pKa = 3.97RR103 pKa = 11.84AHH105 pKa = 6.2CAKK108 pKa = 10.07IKK110 pKa = 10.02YY111 pKa = 9.52FVVTEE116 pKa = 3.98YY117 pKa = 11.6GDD119 pKa = 3.84GKK121 pKa = 10.68GSRR124 pKa = 11.84GKK126 pKa = 10.72GNNPHH131 pKa = 5.87FHH133 pKa = 6.73VIFMFQPHH141 pKa = 6.57ANVGKK146 pKa = 9.82HH147 pKa = 3.7LTGRR151 pKa = 11.84KK152 pKa = 8.77KK153 pKa = 10.62SYY155 pKa = 10.27MNIFFPPTPHH165 pKa = 7.1QMLQYY170 pKa = 9.7MRR172 pKa = 11.84YY173 pKa = 7.91YY174 pKa = 8.84WQGNKK179 pKa = 7.5ITNVYY184 pKa = 10.08HH185 pKa = 7.4DD186 pKa = 3.95GSFDD190 pKa = 3.53VEE192 pKa = 4.96SYY194 pKa = 11.0KK195 pKa = 10.41RR196 pKa = 11.84WQDD199 pKa = 3.19LKK201 pKa = 11.32YY202 pKa = 10.78GFVKK206 pKa = 10.46PGCLKK211 pKa = 10.36GARR214 pKa = 11.84NGAACGEE221 pKa = 4.15VLPDD225 pKa = 4.27NIDD228 pKa = 3.28ALGYY232 pKa = 6.09VTKK235 pKa = 10.88YY236 pKa = 6.8VTKK239 pKa = 8.83DD240 pKa = 2.86THH242 pKa = 5.36KK243 pKa = 10.63RR244 pKa = 11.84RR245 pKa = 11.84VEE247 pKa = 3.75TEE249 pKa = 3.55IYY251 pKa = 9.59RR252 pKa = 11.84RR253 pKa = 11.84IYY255 pKa = 9.63GEE257 pKa = 4.65SYY259 pKa = 10.58FDD261 pKa = 3.94CANNLDD267 pKa = 3.75LVYY270 pKa = 10.44RR271 pKa = 11.84CLRR274 pKa = 11.84FSVEE278 pKa = 4.07EE279 pKa = 4.01KK280 pKa = 10.44NKK282 pKa = 10.05EE283 pKa = 3.84FASEE287 pKa = 4.27YY288 pKa = 10.58EE289 pKa = 4.33DD290 pKa = 3.38IKK292 pKa = 11.57GDD294 pKa = 3.58VNYY297 pKa = 10.77DD298 pKa = 3.78GLSSIYY304 pKa = 10.81DD305 pKa = 3.52NLADD309 pKa = 3.65VGKK312 pKa = 10.95LEE314 pKa = 4.43FSDD317 pKa = 4.93SITEE321 pKa = 4.28KK322 pKa = 10.88LCSAYY327 pKa = 10.59DD328 pKa = 3.79NYY330 pKa = 11.35LHH332 pKa = 6.59SCSSRR337 pKa = 11.84NVSPYY342 pKa = 10.34FFTYY346 pKa = 10.56CMNKK350 pKa = 9.46GVRR353 pKa = 11.84LLFPSLRR360 pKa = 11.84DD361 pKa = 3.5VQLNRR366 pKa = 11.84IKK368 pKa = 10.91HH369 pKa = 5.61LFNKK373 pKa = 8.96VFHH376 pKa = 6.9SMLEE380 pKa = 4.2EE381 pKa = 4.01YY382 pKa = 10.71QNEE385 pKa = 4.23CVKK388 pKa = 10.75YY389 pKa = 9.21YY390 pKa = 10.61RR391 pKa = 11.84NRR393 pKa = 11.84YY394 pKa = 5.1STKK397 pKa = 9.91VRR399 pKa = 11.84ISQHH403 pKa = 5.73FGDD406 pKa = 5.21SIVEE410 pKa = 4.1DD411 pKa = 4.36FRR413 pKa = 11.84EE414 pKa = 3.82KK415 pKa = 10.81ALQNRR420 pKa = 11.84EE421 pKa = 3.97LVYY424 pKa = 11.02LLNINGKK431 pKa = 8.95VNQKK435 pKa = 9.48YY436 pKa = 8.83YY437 pKa = 10.5CGSYY441 pKa = 10.05YY442 pKa = 10.11YY443 pKa = 10.43RR444 pKa = 11.84KK445 pKa = 10.12LFYY448 pKa = 10.61KK449 pKa = 9.88VVKK452 pKa = 10.47GHH454 pKa = 7.06DD455 pKa = 3.56GSNKK459 pKa = 9.7YY460 pKa = 9.86VLNDD464 pKa = 3.44FGIEE468 pKa = 4.01YY469 pKa = 10.74KK470 pKa = 10.25MKK472 pKa = 10.0TLDD475 pKa = 4.61DD476 pKa = 4.52NIEE479 pKa = 4.03RR480 pKa = 11.84QFAKK484 pKa = 10.05YY485 pKa = 10.48KK486 pKa = 10.26RR487 pKa = 11.84MLVSVYY493 pKa = 10.54NDD495 pKa = 3.22CNLYY499 pKa = 9.28STFNDD504 pKa = 2.71WRR506 pKa = 11.84KK507 pKa = 10.52KK508 pKa = 9.69YY509 pKa = 10.47GYY511 pKa = 10.32DD512 pKa = 3.39YY513 pKa = 10.82GISFAGSFFSTLSSIGFSEE532 pKa = 3.88ITEE535 pKa = 4.45DD536 pKa = 4.81DD537 pKa = 4.12LLKK540 pKa = 10.81AAIFKK545 pKa = 10.07IVYY548 pKa = 8.75RR549 pKa = 11.84DD550 pKa = 3.48RR551 pKa = 11.84YY552 pKa = 7.43TDD554 pKa = 3.42CVGINHH560 pKa = 6.01VDD562 pKa = 3.13YY563 pKa = 8.24FTRR566 pKa = 11.84YY567 pKa = 7.76RR568 pKa = 11.84DD569 pKa = 3.62YY570 pKa = 10.65YY571 pKa = 8.6EE572 pKa = 5.3SYY574 pKa = 10.37IIPHH578 pKa = 6.04LVEE581 pKa = 4.85NYY583 pKa = 10.58SSLTSLYY590 pKa = 9.85DD591 pKa = 3.3INMKK595 pKa = 7.73TVCAYY600 pKa = 9.19PLSDD604 pKa = 3.5NVVLSPYY611 pKa = 10.95SKK613 pKa = 10.99LCDD616 pKa = 3.52LFEE619 pKa = 4.83ALNDD623 pKa = 3.8YY624 pKa = 10.33IVYY627 pKa = 10.34LRR629 pKa = 11.84DD630 pKa = 3.62IQWSNDD636 pKa = 3.09YY637 pKa = 10.57EE638 pKa = 4.23EE639 pKa = 4.86RR640 pKa = 11.84SRR642 pKa = 11.84LRR644 pKa = 11.84KK645 pKa = 8.48LHH647 pKa = 6.09KK648 pKa = 9.66RR649 pKa = 11.84SEE651 pKa = 4.13VEE653 pKa = 3.92EE654 pKa = 3.96YY655 pKa = 10.9LVANRR660 pKa = 11.84YY661 pKa = 7.76LL662 pKa = 3.59
Molecular weight: 78.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1703 |
95 |
662 |
425.8 |
48.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.166 ± 1.419 | 1.644 ± 0.531 |
6.166 ± 0.322 | 4.815 ± 0.607 |
4.521 ± 0.448 | 6.459 ± 0.746 |
1.82 ± 0.516 | 5.578 ± 0.324 |
5.872 ± 0.97 | 7.575 ± 0.345 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.642 ± 0.487 | 6.753 ± 0.562 |
3.23 ± 0.627 | 3.699 ± 0.869 |
4.756 ± 0.886 | 8.28 ± 0.711 |
5.52 ± 0.929 | 6.577 ± 0.918 |
0.998 ± 0.203 | 6.929 ± 1.744 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
