
Poophage SC_4_H6H8_2017
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.92
Get precalculated fractions of proteins
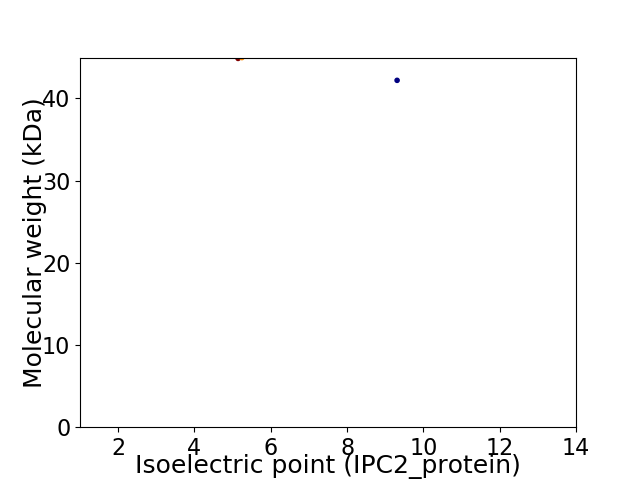
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6H0X6Q9|A0A6H0X6Q9_9VIRU Major capsid protein OS=Poophage SC_4_H6H8_2017 OX=2723248 PE=4 SV=1
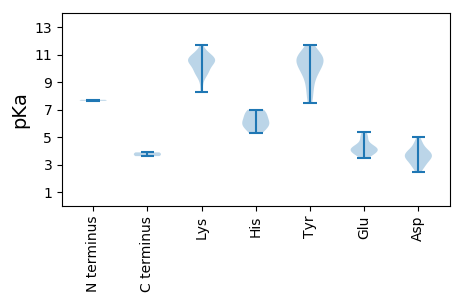
MM1 pKa = 7.71FYY3 pKa = 10.55PRR5 pKa = 11.84DD6 pKa = 3.42RR7 pKa = 11.84KK8 pKa = 10.38ISVDD12 pKa = 3.47RR13 pKa = 11.84DD14 pKa = 3.02EE15 pKa = 4.93LTWNRR20 pKa = 11.84SDD22 pKa = 4.58LFIEE26 pKa = 4.47ILRR29 pKa = 11.84SDD31 pKa = 4.75LFNKK35 pKa = 8.29GTEE38 pKa = 3.81AATPDD43 pKa = 4.0FNKK46 pKa = 10.02LLQMFPQNIDD56 pKa = 3.43VNVPAYY62 pKa = 9.43PYY64 pKa = 10.45DD65 pKa = 3.69VQEE68 pKa = 5.4PKK70 pKa = 11.04VNWNNGEE77 pKa = 4.17GTDD80 pKa = 3.39TSTPSKK86 pKa = 10.89VYY88 pKa = 9.9FAATLNVPFLAAHH101 pKa = 6.71PMAVCPSSPDD111 pKa = 3.03RR112 pKa = 11.84FSRR115 pKa = 11.84LMPPGDD121 pKa = 4.01SNSDD125 pKa = 2.95VDD127 pKa = 4.08FTGVKK132 pKa = 9.41TIPQLAVATRR142 pKa = 11.84LQEE145 pKa = 4.14YY146 pKa = 9.86KK147 pKa = 10.89DD148 pKa = 4.36LIGASGSRR156 pKa = 11.84YY157 pKa = 10.07SDD159 pKa = 2.73WLYY162 pKa = 10.59TFFASKK168 pKa = 9.86IEE170 pKa = 4.19HH171 pKa = 6.01VDD173 pKa = 3.36RR174 pKa = 11.84PKK176 pKa = 11.08LLFSSSVMVNSQVVMNQAGQSGFAGGEE203 pKa = 3.85AAALGQMGGSIAFNSVLGRR222 pKa = 11.84EE223 pKa = 3.68QTYY226 pKa = 8.84YY227 pKa = 10.69FKK229 pKa = 10.98EE230 pKa = 3.83PGYY233 pKa = 10.21IFDD236 pKa = 4.48MLTIRR241 pKa = 11.84PVYY244 pKa = 8.65FWTGIRR250 pKa = 11.84PDD252 pKa = 3.5YY253 pKa = 10.95LEE255 pKa = 4.15YY256 pKa = 10.7RR257 pKa = 11.84GPDD260 pKa = 3.67YY261 pKa = 11.0FNPIYY266 pKa = 10.87NDD268 pKa = 2.44IGYY271 pKa = 9.79QDD273 pKa = 3.41VPFWRR278 pKa = 11.84IGYY281 pKa = 7.85GWKK284 pKa = 10.4GGSPSQSMTVAKK296 pKa = 9.54EE297 pKa = 3.45PCYY300 pKa = 10.87NEE302 pKa = 3.93FRR304 pKa = 11.84SSYY307 pKa = 11.26DD308 pKa = 3.25EE309 pKa = 4.04VLGTLQSTLTPKK321 pKa = 9.87ATVPLQSYY329 pKa = 7.57WVQQRR334 pKa = 11.84DD335 pKa = 4.13FYY337 pKa = 11.56SIGLSSNYY345 pKa = 10.12NEE347 pKa = 5.05ISPSMLFTNLSTVNNPFASDD367 pKa = 3.64LEE369 pKa = 4.92DD370 pKa = 3.32NFFVNMSYY378 pKa = 10.9KK379 pKa = 10.43VVVKK383 pKa = 10.64SLVNKK388 pKa = 10.29SFATRR393 pKa = 11.84LSSRR397 pKa = 3.61
MM1 pKa = 7.71FYY3 pKa = 10.55PRR5 pKa = 11.84DD6 pKa = 3.42RR7 pKa = 11.84KK8 pKa = 10.38ISVDD12 pKa = 3.47RR13 pKa = 11.84DD14 pKa = 3.02EE15 pKa = 4.93LTWNRR20 pKa = 11.84SDD22 pKa = 4.58LFIEE26 pKa = 4.47ILRR29 pKa = 11.84SDD31 pKa = 4.75LFNKK35 pKa = 8.29GTEE38 pKa = 3.81AATPDD43 pKa = 4.0FNKK46 pKa = 10.02LLQMFPQNIDD56 pKa = 3.43VNVPAYY62 pKa = 9.43PYY64 pKa = 10.45DD65 pKa = 3.69VQEE68 pKa = 5.4PKK70 pKa = 11.04VNWNNGEE77 pKa = 4.17GTDD80 pKa = 3.39TSTPSKK86 pKa = 10.89VYY88 pKa = 9.9FAATLNVPFLAAHH101 pKa = 6.71PMAVCPSSPDD111 pKa = 3.03RR112 pKa = 11.84FSRR115 pKa = 11.84LMPPGDD121 pKa = 4.01SNSDD125 pKa = 2.95VDD127 pKa = 4.08FTGVKK132 pKa = 9.41TIPQLAVATRR142 pKa = 11.84LQEE145 pKa = 4.14YY146 pKa = 9.86KK147 pKa = 10.89DD148 pKa = 4.36LIGASGSRR156 pKa = 11.84YY157 pKa = 10.07SDD159 pKa = 2.73WLYY162 pKa = 10.59TFFASKK168 pKa = 9.86IEE170 pKa = 4.19HH171 pKa = 6.01VDD173 pKa = 3.36RR174 pKa = 11.84PKK176 pKa = 11.08LLFSSSVMVNSQVVMNQAGQSGFAGGEE203 pKa = 3.85AAALGQMGGSIAFNSVLGRR222 pKa = 11.84EE223 pKa = 3.68QTYY226 pKa = 8.84YY227 pKa = 10.69FKK229 pKa = 10.98EE230 pKa = 3.83PGYY233 pKa = 10.21IFDD236 pKa = 4.48MLTIRR241 pKa = 11.84PVYY244 pKa = 8.65FWTGIRR250 pKa = 11.84PDD252 pKa = 3.5YY253 pKa = 10.95LEE255 pKa = 4.15YY256 pKa = 10.7RR257 pKa = 11.84GPDD260 pKa = 3.67YY261 pKa = 11.0FNPIYY266 pKa = 10.87NDD268 pKa = 2.44IGYY271 pKa = 9.79QDD273 pKa = 3.41VPFWRR278 pKa = 11.84IGYY281 pKa = 7.85GWKK284 pKa = 10.4GGSPSQSMTVAKK296 pKa = 9.54EE297 pKa = 3.45PCYY300 pKa = 10.87NEE302 pKa = 3.93FRR304 pKa = 11.84SSYY307 pKa = 11.26DD308 pKa = 3.25EE309 pKa = 4.04VLGTLQSTLTPKK321 pKa = 9.87ATVPLQSYY329 pKa = 7.57WVQQRR334 pKa = 11.84DD335 pKa = 4.13FYY337 pKa = 11.56SIGLSSNYY345 pKa = 10.12NEE347 pKa = 5.05ISPSMLFTNLSTVNNPFASDD367 pKa = 3.64LEE369 pKa = 4.92DD370 pKa = 3.32NFFVNMSYY378 pKa = 10.9KK379 pKa = 10.43VVVKK383 pKa = 10.64SLVNKK388 pKa = 10.29SFATRR393 pKa = 11.84LSSRR397 pKa = 3.61
Molecular weight: 44.86 kDa
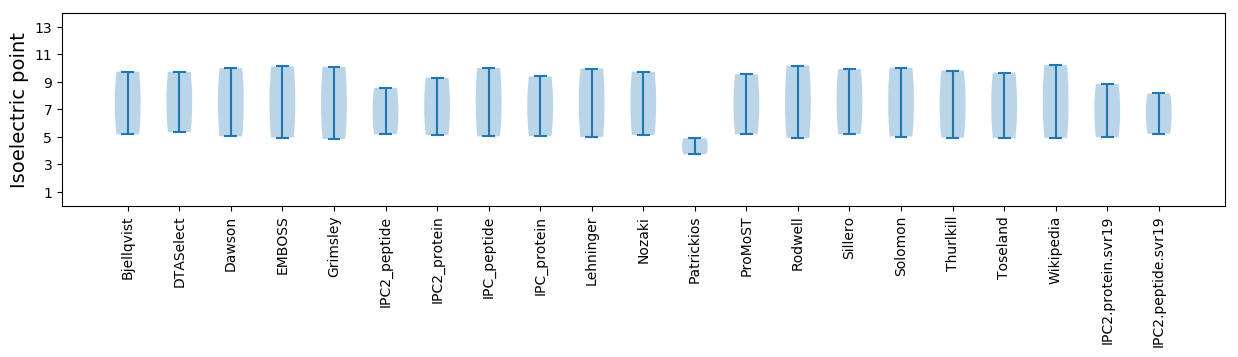
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6H0X6Q9|A0A6H0X6Q9_9VIRU Major capsid protein OS=Poophage SC_4_H6H8_2017 OX=2723248 PE=4 SV=1
MM1 pKa = 7.63CLAPIWIRR9 pKa = 11.84NRR11 pKa = 11.84AYY13 pKa = 10.37SSRR16 pKa = 11.84TIGLTDD22 pKa = 3.81RR23 pKa = 11.84KK24 pKa = 9.77VLLMNRR30 pKa = 11.84PWDD33 pKa = 3.75YY34 pKa = 8.37FTQRR38 pKa = 11.84IMVPCGRR45 pKa = 11.84CEE47 pKa = 3.64EE48 pKa = 4.66CLRR51 pKa = 11.84QQRR54 pKa = 11.84NDD56 pKa = 2.43WYY58 pKa = 10.3IRR60 pKa = 11.84LEE62 pKa = 4.44RR63 pKa = 11.84EE64 pKa = 3.68TKK66 pKa = 8.74YY67 pKa = 10.74QKK69 pKa = 10.45SLYY72 pKa = 8.7RR73 pKa = 11.84NSVFVTITIAPEE85 pKa = 4.25YY86 pKa = 10.13YY87 pKa = 10.48DD88 pKa = 4.65SALQNPSSFIRR99 pKa = 11.84LWFEE103 pKa = 4.13RR104 pKa = 11.84IRR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84FGHH111 pKa = 5.88SIKK114 pKa = 10.55HH115 pKa = 5.69AVFQEE120 pKa = 4.44FGVHH124 pKa = 6.6PEE126 pKa = 4.16LGNEE130 pKa = 4.02PRR132 pKa = 11.84LHH134 pKa = 5.27FHH136 pKa = 6.16GVLWDD141 pKa = 3.78VPHH144 pKa = 6.99SYY146 pKa = 10.8NAIRR150 pKa = 11.84EE151 pKa = 4.29AVKK154 pKa = 10.73DD155 pKa = 4.02LGFVWIASITDD166 pKa = 2.8KK167 pKa = 10.78RR168 pKa = 11.84LRR170 pKa = 11.84YY171 pKa = 8.12VVKK174 pKa = 10.49YY175 pKa = 9.69VGKK178 pKa = 9.73SVYY181 pKa = 9.29MDD183 pKa = 3.17EE184 pKa = 5.22RR185 pKa = 11.84SADD188 pKa = 3.64FAKK191 pKa = 10.58SLPITVGKK199 pKa = 10.67LNTNLYY205 pKa = 11.01DD206 pKa = 3.75FLQNSKK212 pKa = 9.29YY213 pKa = 9.52RR214 pKa = 11.84RR215 pKa = 11.84KK216 pKa = 10.03FISAGVGDD224 pKa = 3.9YY225 pKa = 11.24LGDD228 pKa = 4.28FKK230 pKa = 11.68APGAASGLWSYY241 pKa = 11.63TDD243 pKa = 3.86FKK245 pKa = 10.7TGVVYY250 pKa = 10.05RR251 pKa = 11.84YY252 pKa = 9.64RR253 pKa = 11.84IPRR256 pKa = 11.84YY257 pKa = 7.48YY258 pKa = 10.73DD259 pKa = 3.09KK260 pKa = 11.34YY261 pKa = 10.65LSQDD265 pKa = 2.77ALVFRR270 pKa = 11.84KK271 pKa = 9.92ISTAWTYY278 pKa = 11.7ASAFGGSLALGFLRR292 pKa = 11.84EE293 pKa = 3.85VAEE296 pKa = 4.16RR297 pKa = 11.84VLRR300 pKa = 11.84PSDD303 pKa = 3.52FSRR306 pKa = 11.84VVKK309 pKa = 10.65GGFSRR314 pKa = 11.84LVKK317 pKa = 10.46LRR319 pKa = 11.84EE320 pKa = 3.91FLSKK324 pKa = 11.05VKK326 pKa = 10.43DD327 pKa = 3.6RR328 pKa = 11.84PSFLAVTSDD337 pKa = 5.01VIDD340 pKa = 4.14FWVDD344 pKa = 2.93CFGIDD349 pKa = 3.84PSNPFFNKK357 pKa = 9.48IVYY360 pKa = 9.16GG361 pKa = 3.88
MM1 pKa = 7.63CLAPIWIRR9 pKa = 11.84NRR11 pKa = 11.84AYY13 pKa = 10.37SSRR16 pKa = 11.84TIGLTDD22 pKa = 3.81RR23 pKa = 11.84KK24 pKa = 9.77VLLMNRR30 pKa = 11.84PWDD33 pKa = 3.75YY34 pKa = 8.37FTQRR38 pKa = 11.84IMVPCGRR45 pKa = 11.84CEE47 pKa = 3.64EE48 pKa = 4.66CLRR51 pKa = 11.84QQRR54 pKa = 11.84NDD56 pKa = 2.43WYY58 pKa = 10.3IRR60 pKa = 11.84LEE62 pKa = 4.44RR63 pKa = 11.84EE64 pKa = 3.68TKK66 pKa = 8.74YY67 pKa = 10.74QKK69 pKa = 10.45SLYY72 pKa = 8.7RR73 pKa = 11.84NSVFVTITIAPEE85 pKa = 4.25YY86 pKa = 10.13YY87 pKa = 10.48DD88 pKa = 4.65SALQNPSSFIRR99 pKa = 11.84LWFEE103 pKa = 4.13RR104 pKa = 11.84IRR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84FGHH111 pKa = 5.88SIKK114 pKa = 10.55HH115 pKa = 5.69AVFQEE120 pKa = 4.44FGVHH124 pKa = 6.6PEE126 pKa = 4.16LGNEE130 pKa = 4.02PRR132 pKa = 11.84LHH134 pKa = 5.27FHH136 pKa = 6.16GVLWDD141 pKa = 3.78VPHH144 pKa = 6.99SYY146 pKa = 10.8NAIRR150 pKa = 11.84EE151 pKa = 4.29AVKK154 pKa = 10.73DD155 pKa = 4.02LGFVWIASITDD166 pKa = 2.8KK167 pKa = 10.78RR168 pKa = 11.84LRR170 pKa = 11.84YY171 pKa = 8.12VVKK174 pKa = 10.49YY175 pKa = 9.69VGKK178 pKa = 9.73SVYY181 pKa = 9.29MDD183 pKa = 3.17EE184 pKa = 5.22RR185 pKa = 11.84SADD188 pKa = 3.64FAKK191 pKa = 10.58SLPITVGKK199 pKa = 10.67LNTNLYY205 pKa = 11.01DD206 pKa = 3.75FLQNSKK212 pKa = 9.29YY213 pKa = 9.52RR214 pKa = 11.84RR215 pKa = 11.84KK216 pKa = 10.03FISAGVGDD224 pKa = 3.9YY225 pKa = 11.24LGDD228 pKa = 4.28FKK230 pKa = 11.68APGAASGLWSYY241 pKa = 11.63TDD243 pKa = 3.86FKK245 pKa = 10.7TGVVYY250 pKa = 10.05RR251 pKa = 11.84YY252 pKa = 9.64RR253 pKa = 11.84IPRR256 pKa = 11.84YY257 pKa = 7.48YY258 pKa = 10.73DD259 pKa = 3.09KK260 pKa = 11.34YY261 pKa = 10.65LSQDD265 pKa = 2.77ALVFRR270 pKa = 11.84KK271 pKa = 9.92ISTAWTYY278 pKa = 11.7ASAFGGSLALGFLRR292 pKa = 11.84EE293 pKa = 3.85VAEE296 pKa = 4.16RR297 pKa = 11.84VLRR300 pKa = 11.84PSDD303 pKa = 3.52FSRR306 pKa = 11.84VVKK309 pKa = 10.65GGFSRR314 pKa = 11.84LVKK317 pKa = 10.46LRR319 pKa = 11.84EE320 pKa = 3.91FLSKK324 pKa = 11.05VKK326 pKa = 10.43DD327 pKa = 3.6RR328 pKa = 11.84PSFLAVTSDD337 pKa = 5.01VIDD340 pKa = 4.14FWVDD344 pKa = 2.93CFGIDD349 pKa = 3.84PSNPFFNKK357 pKa = 9.48IVYY360 pKa = 9.16GG361 pKa = 3.88
Molecular weight: 42.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
758 |
361 |
397 |
379.0 |
43.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.673 ± 0.096 | 0.923 ± 0.306 |
6.069 ± 0.166 | 3.958 ± 0.053 |
6.728 ± 0.13 | 6.201 ± 0.07 |
1.055 ± 0.402 | 4.749 ± 0.524 |
4.881 ± 0.62 | 7.784 ± 0.349 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.979 ± 0.577 | 4.749 ± 0.944 |
5.541 ± 0.918 | 3.166 ± 0.629 |
7.124 ± 1.703 | 8.971 ± 0.804 |
4.749 ± 0.577 | 7.652 ± 0.253 |
2.111 ± 0.253 | 5.937 ± 0.104 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
