
Phormidium ambiguum IAM M-71
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Oscillatoriophycideae; Oscillatoriales; Oscillatoriaceae; Phormidium; Phormidium ambiguum
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
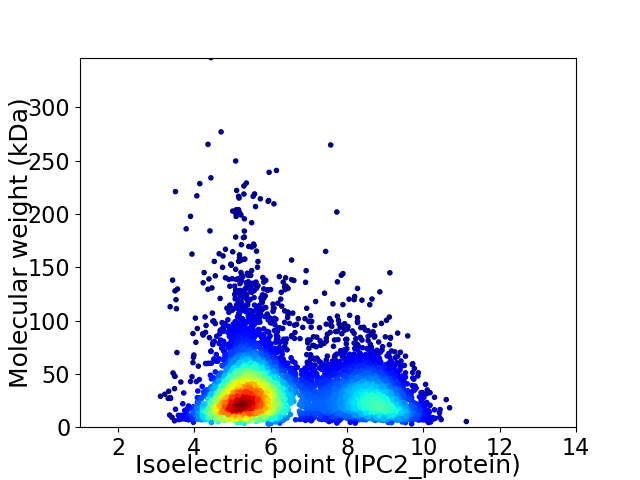
Virtual 2D-PAGE plot for 6148 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1U7I3K8|A0A1U7I3K8_9CYAN Uncharacterized protein OS=Phormidium ambiguum IAM M-71 OX=454136 GN=NIES2119_30420 PE=4 SV=1
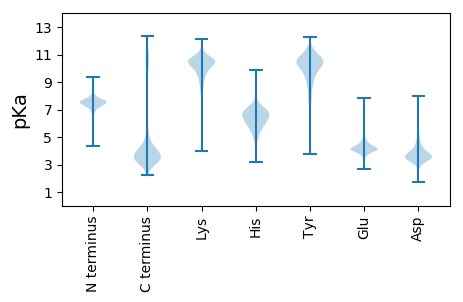
MM1 pKa = 7.16TVNIYY6 pKa = 11.22SNVPGDD12 pKa = 3.71GFASEE17 pKa = 4.12EE18 pKa = 4.15TKK20 pKa = 10.69LYY22 pKa = 11.12NLVNEE27 pKa = 4.19YY28 pKa = 10.22RR29 pKa = 11.84AQNGLAPITASKK41 pKa = 10.77ALSTVANRR49 pKa = 11.84HH50 pKa = 4.81VLDD53 pKa = 3.78LAEE56 pKa = 4.91NIGTLTHH63 pKa = 6.01SWSDD67 pKa = 3.41APYY70 pKa = 10.51DD71 pKa = 3.75PSNSSTYY78 pKa = 10.53SAMWSAPQRR87 pKa = 11.84FNTGYY92 pKa = 10.55VGNGYY97 pKa = 10.31EE98 pKa = 4.1NAFGGSGSYY107 pKa = 9.46IANATDD113 pKa = 5.53ALQAWKK119 pKa = 10.67NSPAHH124 pKa = 5.86NAVILNQGMWQNLQWNALGVGLYY147 pKa = 9.99KK148 pKa = 10.71GYY150 pKa = 11.05AVLWFGQQPDD160 pKa = 3.82TTGTPEE166 pKa = 5.75LITTSSSTSDD176 pKa = 3.36PNTGNNSTSVLASTTPTNSPIFGTEE201 pKa = 3.97GNDD204 pKa = 3.35TLLGTEE210 pKa = 5.01GDD212 pKa = 4.04DD213 pKa = 3.76TLHH216 pKa = 6.75GLGGNDD222 pKa = 3.38LFYY225 pKa = 11.16AGGGNDD231 pKa = 3.16ILFGNLGSDD240 pKa = 3.69SLNGNLGNDD249 pKa = 3.34SLYY252 pKa = 11.0GGKK255 pKa = 9.92DD256 pKa = 3.13ADD258 pKa = 4.1ILWGGKK264 pKa = 10.29GEE266 pKa = 4.12DD267 pKa = 3.47ALYY270 pKa = 11.19GNLGNDD276 pKa = 3.71LLYY279 pKa = 11.33GNIGQDD285 pKa = 2.53ILYY288 pKa = 10.05GGRR291 pKa = 11.84EE292 pKa = 3.95ADD294 pKa = 3.47TLYY297 pKa = 10.97GGQDD301 pKa = 3.57DD302 pKa = 5.32DD303 pKa = 5.27VLFGDD308 pKa = 5.1LDD310 pKa = 3.92NDD312 pKa = 3.61ILFGDD317 pKa = 4.56FGVDD321 pKa = 2.98TLTGGEE327 pKa = 4.11GSDD330 pKa = 3.87IFGLQLDD337 pKa = 4.19RR338 pKa = 11.84GTDD341 pKa = 3.19IVSDD345 pKa = 3.78FTDD348 pKa = 4.09GVDD351 pKa = 4.61FLGLSAGLSFANLSITQGTGTDD373 pKa = 3.38AANTLISANGQLLAVLQGVQAITITEE399 pKa = 3.75ADD401 pKa = 3.17IRR403 pKa = 11.84LLL405 pKa = 3.89
MM1 pKa = 7.16TVNIYY6 pKa = 11.22SNVPGDD12 pKa = 3.71GFASEE17 pKa = 4.12EE18 pKa = 4.15TKK20 pKa = 10.69LYY22 pKa = 11.12NLVNEE27 pKa = 4.19YY28 pKa = 10.22RR29 pKa = 11.84AQNGLAPITASKK41 pKa = 10.77ALSTVANRR49 pKa = 11.84HH50 pKa = 4.81VLDD53 pKa = 3.78LAEE56 pKa = 4.91NIGTLTHH63 pKa = 6.01SWSDD67 pKa = 3.41APYY70 pKa = 10.51DD71 pKa = 3.75PSNSSTYY78 pKa = 10.53SAMWSAPQRR87 pKa = 11.84FNTGYY92 pKa = 10.55VGNGYY97 pKa = 10.31EE98 pKa = 4.1NAFGGSGSYY107 pKa = 9.46IANATDD113 pKa = 5.53ALQAWKK119 pKa = 10.67NSPAHH124 pKa = 5.86NAVILNQGMWQNLQWNALGVGLYY147 pKa = 9.99KK148 pKa = 10.71GYY150 pKa = 11.05AVLWFGQQPDD160 pKa = 3.82TTGTPEE166 pKa = 5.75LITTSSSTSDD176 pKa = 3.36PNTGNNSTSVLASTTPTNSPIFGTEE201 pKa = 3.97GNDD204 pKa = 3.35TLLGTEE210 pKa = 5.01GDD212 pKa = 4.04DD213 pKa = 3.76TLHH216 pKa = 6.75GLGGNDD222 pKa = 3.38LFYY225 pKa = 11.16AGGGNDD231 pKa = 3.16ILFGNLGSDD240 pKa = 3.69SLNGNLGNDD249 pKa = 3.34SLYY252 pKa = 11.0GGKK255 pKa = 9.92DD256 pKa = 3.13ADD258 pKa = 4.1ILWGGKK264 pKa = 10.29GEE266 pKa = 4.12DD267 pKa = 3.47ALYY270 pKa = 11.19GNLGNDD276 pKa = 3.71LLYY279 pKa = 11.33GNIGQDD285 pKa = 2.53ILYY288 pKa = 10.05GGRR291 pKa = 11.84EE292 pKa = 3.95ADD294 pKa = 3.47TLYY297 pKa = 10.97GGQDD301 pKa = 3.57DD302 pKa = 5.32DD303 pKa = 5.27VLFGDD308 pKa = 5.1LDD310 pKa = 3.92NDD312 pKa = 3.61ILFGDD317 pKa = 4.56FGVDD321 pKa = 2.98TLTGGEE327 pKa = 4.11GSDD330 pKa = 3.87IFGLQLDD337 pKa = 4.19RR338 pKa = 11.84GTDD341 pKa = 3.19IVSDD345 pKa = 3.78FTDD348 pKa = 4.09GVDD351 pKa = 4.61FLGLSAGLSFANLSITQGTGTDD373 pKa = 3.38AANTLISANGQLLAVLQGVQAITITEE399 pKa = 3.75ADD401 pKa = 3.17IRR403 pKa = 11.84LLL405 pKa = 3.89
Molecular weight: 42.36 kDa
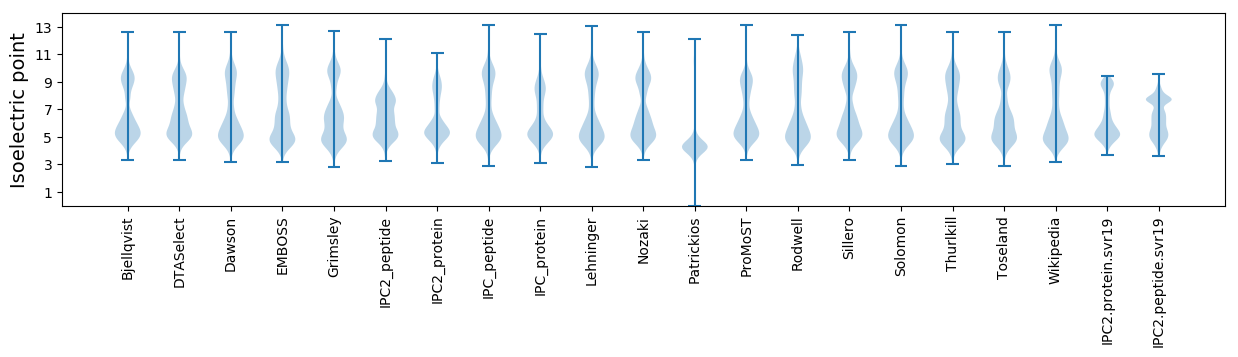
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1U7IJJ4|A0A1U7IJJ4_9CYAN RND transporter OS=Phormidium ambiguum IAM M-71 OX=454136 GN=NIES2119_14070 PE=3 SV=1
MM1 pKa = 6.63TQRR4 pKa = 11.84TLHH7 pKa = 4.82GTSRR11 pKa = 11.84KK12 pKa = 9.46RR13 pKa = 11.84KK14 pKa = 7.05RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TKK26 pKa = 10.45NGKK29 pKa = 9.01LVLKK33 pKa = 10.49ARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.34KK38 pKa = 10.23GRR40 pKa = 11.84YY41 pKa = 8.55RR42 pKa = 11.84LAVV45 pKa = 3.33
MM1 pKa = 6.63TQRR4 pKa = 11.84TLHH7 pKa = 4.82GTSRR11 pKa = 11.84KK12 pKa = 9.46RR13 pKa = 11.84KK14 pKa = 7.05RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TKK26 pKa = 10.45NGKK29 pKa = 9.01LVLKK33 pKa = 10.49ARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.34KK38 pKa = 10.23GRR40 pKa = 11.84YY41 pKa = 8.55RR42 pKa = 11.84LAVV45 pKa = 3.33
Molecular weight: 5.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2062752 |
29 |
3377 |
335.5 |
37.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.855 ± 0.034 | 0.963 ± 0.011 |
4.74 ± 0.029 | 6.588 ± 0.033 |
4.061 ± 0.02 | 6.528 ± 0.039 |
1.664 ± 0.015 | 7.126 ± 0.026 |
5.185 ± 0.029 | 10.924 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.752 ± 0.014 | 4.825 ± 0.037 |
4.614 ± 0.024 | 5.3 ± 0.027 |
4.996 ± 0.024 | 6.365 ± 0.031 |
5.684 ± 0.027 | 6.362 ± 0.026 |
1.449 ± 0.014 | 3.018 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
