
Rhodovulum sp. 12E13
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Rhodovulum; unclassified Rhodovulum
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
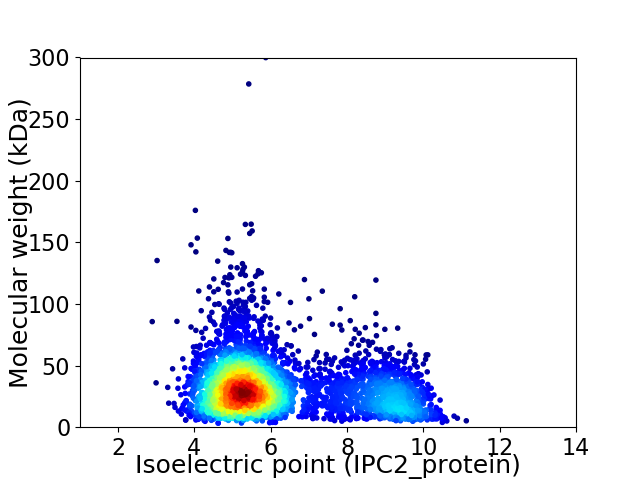
Virtual 2D-PAGE plot for 4102 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A369QZL9|A0A369QZL9_9RHOB Uncharacterized protein OS=Rhodovulum sp. 12E13 OX=2203891 GN=DLJ49_08750 PE=4 SV=1
MM1 pKa = 7.8LFRR4 pKa = 11.84IAKK7 pKa = 9.25AATSAVALIAAVPASAQDD25 pKa = 3.5QEE27 pKa = 4.3QDD29 pKa = 3.64GVEE32 pKa = 4.25LLSNWSYY39 pKa = 11.89DD40 pKa = 3.49PLYY43 pKa = 11.15AEE45 pKa = 4.81GWSVEE50 pKa = 3.8NMFDD54 pKa = 3.02MTEE57 pKa = 3.9IVDD60 pKa = 3.69AAGEE64 pKa = 4.15EE65 pKa = 4.15IGDD68 pKa = 3.74VEE70 pKa = 4.69NVLFANDD77 pKa = 4.22GEE79 pKa = 4.52VLGIIAQVGGFWDD92 pKa = 3.23ILDD95 pKa = 3.63THH97 pKa = 6.86VYY99 pKa = 9.79VPWDD103 pKa = 4.02EE104 pKa = 4.23VTMEE108 pKa = 4.49AGIQRR113 pKa = 11.84MGIPVTEE120 pKa = 4.25EE121 pKa = 3.7TVDD124 pKa = 3.81DD125 pKa = 4.21YY126 pKa = 11.94DD127 pKa = 4.11VFGDD131 pKa = 4.02FDD133 pKa = 4.72FDD135 pKa = 3.62EE136 pKa = 4.3QVIDD140 pKa = 4.21EE141 pKa = 4.68SDD143 pKa = 3.23TGEE146 pKa = 4.19TQPVDD151 pKa = 3.66DD152 pKa = 5.54DD153 pKa = 4.16LAAGAGVFKK162 pKa = 10.58ATDD165 pKa = 3.52LVGDD169 pKa = 3.71YY170 pKa = 10.91AYY172 pKa = 10.93LSDD175 pKa = 3.72GARR178 pKa = 11.84YY179 pKa = 9.75GYY181 pKa = 10.48VADD184 pKa = 6.49LIVQDD189 pKa = 4.14GAISAVVTDD198 pKa = 4.2AAAYY202 pKa = 7.96GRR204 pKa = 11.84PGYY207 pKa = 9.6YY208 pKa = 10.06AYY210 pKa = 9.72PYY212 pKa = 9.63SYY214 pKa = 10.0RR215 pKa = 11.84GAMSPRR221 pKa = 11.84YY222 pKa = 8.35EE223 pKa = 3.93IPYY226 pKa = 10.48DD227 pKa = 3.49PVEE230 pKa = 4.94IDD232 pKa = 3.07TLEE235 pKa = 4.27NFDD238 pKa = 4.23YY239 pKa = 11.15EE240 pKa = 4.38QLQSRR245 pKa = 11.84AMPWRR250 pKa = 11.84SATSPGRR257 pKa = 11.84RR258 pKa = 11.84PSSCAHH264 pKa = 6.13GPAVGLLSGTKK275 pKa = 6.76TTSGRR280 pKa = 11.84RR281 pKa = 11.84LAGQGGCPAPYY292 pKa = 10.0NPCEE296 pKa = 4.64DD297 pKa = 3.7GGAHH301 pKa = 6.58HH302 pKa = 6.89EE303 pKa = 4.2WAVLLGEE310 pKa = 4.25RR311 pKa = 11.84TSPRR315 pKa = 11.84RR316 pKa = 11.84VRR318 pKa = 3.66
MM1 pKa = 7.8LFRR4 pKa = 11.84IAKK7 pKa = 9.25AATSAVALIAAVPASAQDD25 pKa = 3.5QEE27 pKa = 4.3QDD29 pKa = 3.64GVEE32 pKa = 4.25LLSNWSYY39 pKa = 11.89DD40 pKa = 3.49PLYY43 pKa = 11.15AEE45 pKa = 4.81GWSVEE50 pKa = 3.8NMFDD54 pKa = 3.02MTEE57 pKa = 3.9IVDD60 pKa = 3.69AAGEE64 pKa = 4.15EE65 pKa = 4.15IGDD68 pKa = 3.74VEE70 pKa = 4.69NVLFANDD77 pKa = 4.22GEE79 pKa = 4.52VLGIIAQVGGFWDD92 pKa = 3.23ILDD95 pKa = 3.63THH97 pKa = 6.86VYY99 pKa = 9.79VPWDD103 pKa = 4.02EE104 pKa = 4.23VTMEE108 pKa = 4.49AGIQRR113 pKa = 11.84MGIPVTEE120 pKa = 4.25EE121 pKa = 3.7TVDD124 pKa = 3.81DD125 pKa = 4.21YY126 pKa = 11.94DD127 pKa = 4.11VFGDD131 pKa = 4.02FDD133 pKa = 4.72FDD135 pKa = 3.62EE136 pKa = 4.3QVIDD140 pKa = 4.21EE141 pKa = 4.68SDD143 pKa = 3.23TGEE146 pKa = 4.19TQPVDD151 pKa = 3.66DD152 pKa = 5.54DD153 pKa = 4.16LAAGAGVFKK162 pKa = 10.58ATDD165 pKa = 3.52LVGDD169 pKa = 3.71YY170 pKa = 10.91AYY172 pKa = 10.93LSDD175 pKa = 3.72GARR178 pKa = 11.84YY179 pKa = 9.75GYY181 pKa = 10.48VADD184 pKa = 6.49LIVQDD189 pKa = 4.14GAISAVVTDD198 pKa = 4.2AAAYY202 pKa = 7.96GRR204 pKa = 11.84PGYY207 pKa = 9.6YY208 pKa = 10.06AYY210 pKa = 9.72PYY212 pKa = 9.63SYY214 pKa = 10.0RR215 pKa = 11.84GAMSPRR221 pKa = 11.84YY222 pKa = 8.35EE223 pKa = 3.93IPYY226 pKa = 10.48DD227 pKa = 3.49PVEE230 pKa = 4.94IDD232 pKa = 3.07TLEE235 pKa = 4.27NFDD238 pKa = 4.23YY239 pKa = 11.15EE240 pKa = 4.38QLQSRR245 pKa = 11.84AMPWRR250 pKa = 11.84SATSPGRR257 pKa = 11.84RR258 pKa = 11.84PSSCAHH264 pKa = 6.13GPAVGLLSGTKK275 pKa = 6.76TTSGRR280 pKa = 11.84RR281 pKa = 11.84LAGQGGCPAPYY292 pKa = 10.0NPCEE296 pKa = 4.64DD297 pKa = 3.7GGAHH301 pKa = 6.58HH302 pKa = 6.89EE303 pKa = 4.2WAVLLGEE310 pKa = 4.25RR311 pKa = 11.84TSPRR315 pKa = 11.84RR316 pKa = 11.84VRR318 pKa = 3.66
Molecular weight: 34.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A369QR66|A0A369QR66_9RHOB Uncharacterized protein OS=Rhodovulum sp. 12E13 OX=2203891 GN=DLJ49_20180 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1297611 |
31 |
2794 |
316.3 |
33.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.702 ± 0.07 | 0.829 ± 0.012 |
5.841 ± 0.029 | 6.286 ± 0.034 |
3.373 ± 0.025 | 9.39 ± 0.037 |
2.031 ± 0.02 | 4.206 ± 0.03 |
1.966 ± 0.029 | 10.157 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.43 ± 0.019 | 1.835 ± 0.018 |
5.798 ± 0.032 | 2.626 ± 0.019 |
8.082 ± 0.046 | 4.374 ± 0.024 |
5.195 ± 0.026 | 7.503 ± 0.035 |
1.448 ± 0.017 | 1.93 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
