
Lynx canadensis faeces associated genomovirus CL1 148
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus lynca4
Average proteome isoelectric point is 7.46
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
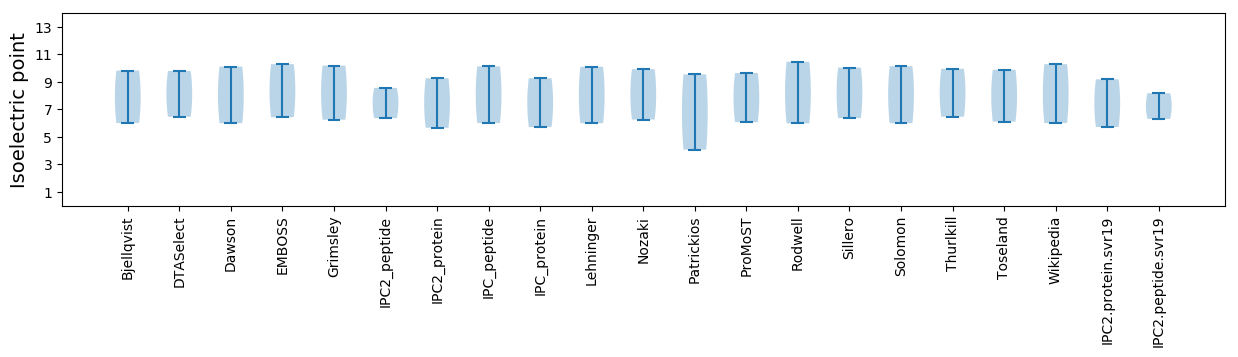
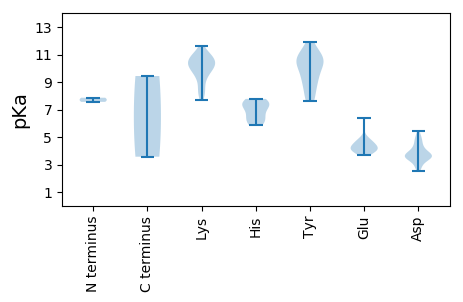
Protein with the lowest isoelectric point:
>tr|A0A2Z5CJ94|A0A2Z5CJ94_9VIRU Replication-associated protein OS=Lynx canadensis faeces associated genomovirus CL1 148 OX=2219123 PE=3 SV=1
MM1 pKa = 7.56SFHH4 pKa = 7.26CNAQYY9 pKa = 11.23FLVTYY14 pKa = 7.6AQCGDD19 pKa = 4.0LDD21 pKa = 3.71PWSVMEE27 pKa = 4.75RR28 pKa = 11.84FSTLGAEE35 pKa = 4.5CIIARR40 pKa = 11.84EE41 pKa = 3.96LHH43 pKa = 6.59ADD45 pKa = 3.73LGLHH49 pKa = 5.87LHH51 pKa = 6.5CFVDD55 pKa = 4.63FGRR58 pKa = 11.84KK59 pKa = 8.02FRR61 pKa = 11.84SRR63 pKa = 11.84KK64 pKa = 7.68TDD66 pKa = 2.79IFDD69 pKa = 3.68VGSSHH74 pKa = 7.51PNIEE78 pKa = 4.23PSRR81 pKa = 11.84GTPWAGYY88 pKa = 10.35DD89 pKa = 3.62YY90 pKa = 10.72AIKK93 pKa = 10.89DD94 pKa = 3.47GDD96 pKa = 4.3VVCGGLARR104 pKa = 11.84PRR106 pKa = 11.84QPRR109 pKa = 11.84SKK111 pKa = 9.31STRR114 pKa = 11.84NDD116 pKa = 2.56VHH118 pKa = 7.69QYY120 pKa = 9.72TEE122 pKa = 3.84ITNADD127 pKa = 4.18DD128 pKa = 5.43IEE130 pKa = 4.78HH131 pKa = 6.04FWEE134 pKa = 4.74LLHH137 pKa = 7.77HH138 pKa = 7.26LDD140 pKa = 3.99PKK142 pKa = 8.83TAVVNFVSATKK153 pKa = 9.94YY154 pKa = 10.69AEE156 pKa = 3.91ARR158 pKa = 11.84FADD161 pKa = 4.05VEE163 pKa = 4.22PGYY166 pKa = 10.37EE167 pKa = 3.7PRR169 pKa = 11.84RR170 pKa = 11.84RR171 pKa = 11.84QDD173 pKa = 3.51FVAGDD178 pKa = 3.71DD179 pKa = 4.01DD180 pKa = 4.95GRR182 pKa = 11.84DD183 pKa = 3.48AVKK186 pKa = 10.75SLVLFGEE193 pKa = 4.38PLTGKK198 pKa = 8.47TDD200 pKa = 3.28WARR203 pKa = 11.84CLGNHH208 pKa = 6.21VYY210 pKa = 10.71TMGIVSGDD218 pKa = 3.57QMLKK222 pKa = 9.94IANPDD227 pKa = 2.76VKK229 pKa = 10.96YY230 pKa = 10.96AVFDD234 pKa = 4.95DD235 pKa = 3.28IRR237 pKa = 11.84GGIKK241 pKa = 9.99FFPSFKK247 pKa = 10.33EE248 pKa = 3.72WLGCQAYY255 pKa = 7.88VTVTKK260 pKa = 10.25KK261 pKa = 10.8YY262 pKa = 10.52KK263 pKa = 9.98EE264 pKa = 4.1PKK266 pKa = 8.34LMKK269 pKa = 9.51WGRR272 pKa = 11.84PSIWLSNKK280 pKa = 9.96DD281 pKa = 3.78PRR283 pKa = 11.84TEE285 pKa = 4.05MDD287 pKa = 3.7PSDD290 pKa = 4.75ANWLEE295 pKa = 4.1ANCTFIEE302 pKa = 4.15INTPIFHH309 pKa = 7.44ANTT312 pKa = 3.57
MM1 pKa = 7.56SFHH4 pKa = 7.26CNAQYY9 pKa = 11.23FLVTYY14 pKa = 7.6AQCGDD19 pKa = 4.0LDD21 pKa = 3.71PWSVMEE27 pKa = 4.75RR28 pKa = 11.84FSTLGAEE35 pKa = 4.5CIIARR40 pKa = 11.84EE41 pKa = 3.96LHH43 pKa = 6.59ADD45 pKa = 3.73LGLHH49 pKa = 5.87LHH51 pKa = 6.5CFVDD55 pKa = 4.63FGRR58 pKa = 11.84KK59 pKa = 8.02FRR61 pKa = 11.84SRR63 pKa = 11.84KK64 pKa = 7.68TDD66 pKa = 2.79IFDD69 pKa = 3.68VGSSHH74 pKa = 7.51PNIEE78 pKa = 4.23PSRR81 pKa = 11.84GTPWAGYY88 pKa = 10.35DD89 pKa = 3.62YY90 pKa = 10.72AIKK93 pKa = 10.89DD94 pKa = 3.47GDD96 pKa = 4.3VVCGGLARR104 pKa = 11.84PRR106 pKa = 11.84QPRR109 pKa = 11.84SKK111 pKa = 9.31STRR114 pKa = 11.84NDD116 pKa = 2.56VHH118 pKa = 7.69QYY120 pKa = 9.72TEE122 pKa = 3.84ITNADD127 pKa = 4.18DD128 pKa = 5.43IEE130 pKa = 4.78HH131 pKa = 6.04FWEE134 pKa = 4.74LLHH137 pKa = 7.77HH138 pKa = 7.26LDD140 pKa = 3.99PKK142 pKa = 8.83TAVVNFVSATKK153 pKa = 9.94YY154 pKa = 10.69AEE156 pKa = 3.91ARR158 pKa = 11.84FADD161 pKa = 4.05VEE163 pKa = 4.22PGYY166 pKa = 10.37EE167 pKa = 3.7PRR169 pKa = 11.84RR170 pKa = 11.84RR171 pKa = 11.84QDD173 pKa = 3.51FVAGDD178 pKa = 3.71DD179 pKa = 4.01DD180 pKa = 4.95GRR182 pKa = 11.84DD183 pKa = 3.48AVKK186 pKa = 10.75SLVLFGEE193 pKa = 4.38PLTGKK198 pKa = 8.47TDD200 pKa = 3.28WARR203 pKa = 11.84CLGNHH208 pKa = 6.21VYY210 pKa = 10.71TMGIVSGDD218 pKa = 3.57QMLKK222 pKa = 9.94IANPDD227 pKa = 2.76VKK229 pKa = 10.96YY230 pKa = 10.96AVFDD234 pKa = 4.95DD235 pKa = 3.28IRR237 pKa = 11.84GGIKK241 pKa = 9.99FFPSFKK247 pKa = 10.33EE248 pKa = 3.72WLGCQAYY255 pKa = 7.88VTVTKK260 pKa = 10.25KK261 pKa = 10.8YY262 pKa = 10.52KK263 pKa = 9.98EE264 pKa = 4.1PKK266 pKa = 8.34LMKK269 pKa = 9.51WGRR272 pKa = 11.84PSIWLSNKK280 pKa = 9.96DD281 pKa = 3.78PRR283 pKa = 11.84TEE285 pKa = 4.05MDD287 pKa = 3.7PSDD290 pKa = 4.75ANWLEE295 pKa = 4.1ANCTFIEE302 pKa = 4.15INTPIFHH309 pKa = 7.44ANTT312 pKa = 3.57
Molecular weight: 35.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5CJ94|A0A2Z5CJ94_9VIRU Replication-associated protein OS=Lynx canadensis faeces associated genomovirus CL1 148 OX=2219123 PE=3 SV=1
MM1 pKa = 7.86PYY3 pKa = 8.52STKK6 pKa = 10.18RR7 pKa = 11.84GSRR10 pKa = 11.84RR11 pKa = 11.84SYY13 pKa = 8.95TKK15 pKa = 9.97RR16 pKa = 11.84GGFKK20 pKa = 10.39RR21 pKa = 11.84RR22 pKa = 11.84TAPKK26 pKa = 9.54RR27 pKa = 11.84RR28 pKa = 11.84TSYY31 pKa = 10.1RR32 pKa = 11.84RR33 pKa = 11.84SRR35 pKa = 11.84YY36 pKa = 9.27SRR38 pKa = 11.84PKK40 pKa = 8.31RR41 pKa = 11.84TVMPKK46 pKa = 10.25KK47 pKa = 10.42RR48 pKa = 11.84ILNLTSTKK56 pKa = 10.2KK57 pKa = 10.43RR58 pKa = 11.84NTMLQYY64 pKa = 11.51ANTSTSGGGSVTIGPGPYY82 pKa = 10.0LVTGALGGFSIFCPTAMDD100 pKa = 3.69LTEE103 pKa = 5.03KK104 pKa = 10.62SGANNSVVNVAEE116 pKa = 4.51RR117 pKa = 11.84NSTTCFMRR125 pKa = 11.84GFSEE129 pKa = 4.03NLRR132 pKa = 11.84IQTSTGIPWFWRR144 pKa = 11.84RR145 pKa = 11.84ITFCSKK151 pKa = 10.42SGVFGVFDD159 pKa = 4.21SGDD162 pKa = 3.5TPTQTNAGEE171 pKa = 4.1TSYY174 pKa = 11.88VDD176 pKa = 3.32TTNGMEE182 pKa = 4.48RR183 pKa = 11.84LWFNQNVNSAPATVTSIQAIMFKK206 pKa = 10.66GQYY209 pKa = 9.22NKK211 pKa = 10.95DD212 pKa = 3.03WVDD215 pKa = 3.5PLTAPLDD222 pKa = 3.65TARR225 pKa = 11.84IDD227 pKa = 3.69VKK229 pKa = 11.25SDD231 pKa = 2.94TTKK234 pKa = 10.86CIRR237 pKa = 11.84SGNASGTVFEE247 pKa = 4.74KK248 pKa = 10.73KK249 pKa = 9.21CWYY252 pKa = 9.53PMNKK256 pKa = 9.47NLVYY260 pKa = 10.83DD261 pKa = 4.46DD262 pKa = 5.1DD263 pKa = 4.63EE264 pKa = 6.36SGEE267 pKa = 4.47GEE269 pKa = 4.17VARR272 pKa = 11.84YY273 pKa = 7.82TSVVDD278 pKa = 3.53KK279 pKa = 11.06KK280 pKa = 11.64GMGDD284 pKa = 3.57YY285 pKa = 10.55FIVDD289 pKa = 3.52IFVVGTGGATADD301 pKa = 3.56QLQVTGTASMYY312 pKa = 8.7WHH314 pKa = 7.06EE315 pKa = 4.33KK316 pKa = 9.44
MM1 pKa = 7.86PYY3 pKa = 8.52STKK6 pKa = 10.18RR7 pKa = 11.84GSRR10 pKa = 11.84RR11 pKa = 11.84SYY13 pKa = 8.95TKK15 pKa = 9.97RR16 pKa = 11.84GGFKK20 pKa = 10.39RR21 pKa = 11.84RR22 pKa = 11.84TAPKK26 pKa = 9.54RR27 pKa = 11.84RR28 pKa = 11.84TSYY31 pKa = 10.1RR32 pKa = 11.84RR33 pKa = 11.84SRR35 pKa = 11.84YY36 pKa = 9.27SRR38 pKa = 11.84PKK40 pKa = 8.31RR41 pKa = 11.84TVMPKK46 pKa = 10.25KK47 pKa = 10.42RR48 pKa = 11.84ILNLTSTKK56 pKa = 10.2KK57 pKa = 10.43RR58 pKa = 11.84NTMLQYY64 pKa = 11.51ANTSTSGGGSVTIGPGPYY82 pKa = 10.0LVTGALGGFSIFCPTAMDD100 pKa = 3.69LTEE103 pKa = 5.03KK104 pKa = 10.62SGANNSVVNVAEE116 pKa = 4.51RR117 pKa = 11.84NSTTCFMRR125 pKa = 11.84GFSEE129 pKa = 4.03NLRR132 pKa = 11.84IQTSTGIPWFWRR144 pKa = 11.84RR145 pKa = 11.84ITFCSKK151 pKa = 10.42SGVFGVFDD159 pKa = 4.21SGDD162 pKa = 3.5TPTQTNAGEE171 pKa = 4.1TSYY174 pKa = 11.88VDD176 pKa = 3.32TTNGMEE182 pKa = 4.48RR183 pKa = 11.84LWFNQNVNSAPATVTSIQAIMFKK206 pKa = 10.66GQYY209 pKa = 9.22NKK211 pKa = 10.95DD212 pKa = 3.03WVDD215 pKa = 3.5PLTAPLDD222 pKa = 3.65TARR225 pKa = 11.84IDD227 pKa = 3.69VKK229 pKa = 11.25SDD231 pKa = 2.94TTKK234 pKa = 10.86CIRR237 pKa = 11.84SGNASGTVFEE247 pKa = 4.74KK248 pKa = 10.73KK249 pKa = 9.21CWYY252 pKa = 9.53PMNKK256 pKa = 9.47NLVYY260 pKa = 10.83DD261 pKa = 4.46DD262 pKa = 5.1DD263 pKa = 4.63EE264 pKa = 6.36SGEE267 pKa = 4.47GEE269 pKa = 4.17VARR272 pKa = 11.84YY273 pKa = 7.82TSVVDD278 pKa = 3.53KK279 pKa = 11.06KK280 pKa = 11.64GMGDD284 pKa = 3.57YY285 pKa = 10.55FIVDD289 pKa = 3.52IFVVGTGGATADD301 pKa = 3.56QLQVTGTASMYY312 pKa = 8.7WHH314 pKa = 7.06EE315 pKa = 4.33KK316 pKa = 9.44
Molecular weight: 35.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
628 |
312 |
316 |
314.0 |
35.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.369 ± 0.775 | 2.07 ± 0.382 |
6.847 ± 1.396 | 4.14 ± 0.764 |
5.096 ± 0.521 | 8.28 ± 0.95 |
1.911 ± 1.248 | 4.299 ± 0.393 |
6.21 ± 0.341 | 5.096 ± 1.016 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.548 ± 0.483 | 4.618 ± 0.596 |
4.777 ± 0.519 | 2.389 ± 0.112 |
6.847 ± 0.585 | 7.006 ± 1.451 |
8.758 ± 2.31 | 6.688 ± 0.215 |
2.229 ± 0.259 | 3.822 ± 0.229 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
