
Capybara microvirus Cap1_SP_64
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.24
Get precalculated fractions of proteins
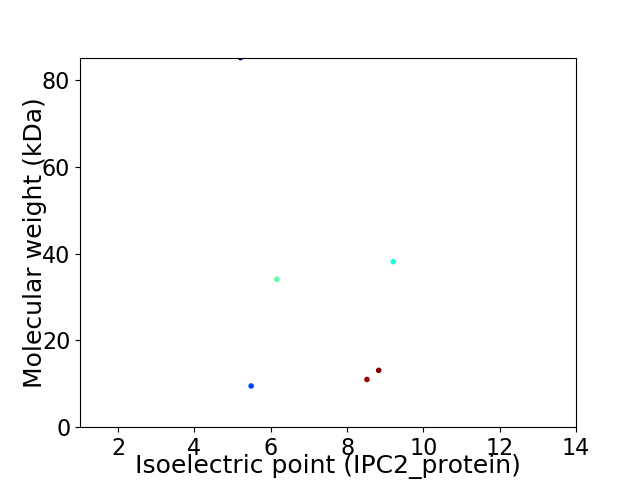
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W7J2|A0A4P8W7J2_9VIRU Replication initiator protein OS=Capybara microvirus Cap1_SP_64 OX=2584787 PE=4 SV=1
MM1 pKa = 7.63AIEE4 pKa = 4.05NSFVYY9 pKa = 10.62SLTPVDD15 pKa = 3.31QTDD18 pKa = 3.56YY19 pKa = 11.43NKK21 pKa = 10.22FDD23 pKa = 3.8FGYY26 pKa = 10.87GKK28 pKa = 10.64YY29 pKa = 8.31FTAKK33 pKa = 9.58PGKK36 pKa = 9.68FYY38 pKa = 10.23PLHH41 pKa = 5.91WEE43 pKa = 4.22EE44 pKa = 4.88IEE46 pKa = 4.23PGANIHH52 pKa = 6.93SKK54 pKa = 9.13IQHH57 pKa = 6.04FARR60 pKa = 11.84TQPLVAPSMIDD71 pKa = 3.23MNIEE75 pKa = 3.98TFSFLVPFRR84 pKa = 11.84IIFDD88 pKa = 3.76GWQEE92 pKa = 4.11FISFGDD98 pKa = 3.56GKK100 pKa = 8.06TLKK103 pKa = 10.25NDD105 pKa = 3.74LGINSQIPYY114 pKa = 8.78ITTQQLLQYY123 pKa = 9.17FAHH126 pKa = 7.33DD127 pKa = 5.35LSCPWNLGFKK137 pKa = 10.01PDD139 pKa = 3.23NCNYY143 pKa = 9.49YY144 pKa = 8.69VTYY147 pKa = 10.47KK148 pKa = 10.77DD149 pKa = 5.65LYY151 pKa = 9.21QCCEE155 pKa = 4.04LLDD158 pKa = 3.92IPMSNLIDD166 pKa = 3.44IDD168 pKa = 3.93EE169 pKa = 4.55FQSYY173 pKa = 8.2DD174 pKa = 3.62TIKK177 pKa = 10.18TFILQTFTHH186 pKa = 7.05FSLTDD191 pKa = 3.13TGQKK195 pKa = 8.81FTIDD199 pKa = 3.35QLFTSQYY206 pKa = 9.64VQIFDD211 pKa = 4.72DD212 pKa = 4.57LNIHH216 pKa = 6.24TFPVNPQNFATLHH229 pKa = 5.86QITNSASTNLAFPKK243 pKa = 10.59YY244 pKa = 9.47SFSKK248 pKa = 10.54KK249 pKa = 10.09SIWYY253 pKa = 8.33NALNLYY259 pKa = 9.51HH260 pKa = 7.11DD261 pKa = 4.78KK262 pKa = 10.71EE263 pKa = 4.56YY264 pKa = 10.9EE265 pKa = 4.07SHH267 pKa = 7.14DD268 pKa = 3.48TQLYY272 pKa = 8.26YY273 pKa = 10.85DD274 pKa = 3.82NKK276 pKa = 8.6EE277 pKa = 4.34TYY279 pKa = 7.69DD280 pKa = 3.66TSSSSLWRR288 pKa = 11.84TRR290 pKa = 11.84LLNKK294 pKa = 10.45LEE296 pKa = 4.27TNNIRR301 pKa = 11.84SYY303 pKa = 10.18WLYY306 pKa = 11.17KK307 pKa = 10.77EE308 pKa = 5.64GDD310 pKa = 4.02TKK312 pKa = 11.01PSALWFATTPQIKK325 pKa = 8.37TLNNSFKK332 pKa = 10.59FAQSKK337 pKa = 10.69INLLPFFAKK346 pKa = 10.16QKK348 pKa = 10.63VYY350 pKa = 10.83EE351 pKa = 4.09DD352 pKa = 3.43WFRR355 pKa = 11.84CEE357 pKa = 3.97PFEE360 pKa = 4.43TEE362 pKa = 3.84YY363 pKa = 11.26SEE365 pKa = 4.01YY366 pKa = 10.5TIFSGKK372 pKa = 7.53VTEE375 pKa = 4.76LPTSEE380 pKa = 4.11FVKK383 pKa = 10.67VLHH386 pKa = 6.76LSPAHH391 pKa = 5.58YY392 pKa = 10.25KK393 pKa = 9.97RR394 pKa = 11.84DD395 pKa = 3.69YY396 pKa = 10.48FQAALPQPTALQQVFVPFSSPIDD419 pKa = 3.52LPGISNPFSDD429 pKa = 4.28NRR431 pKa = 11.84PVTLQMNSDD440 pKa = 3.15KK441 pKa = 11.13TLFAEE446 pKa = 4.55YY447 pKa = 10.23KK448 pKa = 9.7PDD450 pKa = 3.57PTRR453 pKa = 11.84STIAEE458 pKa = 3.96RR459 pKa = 11.84VEE461 pKa = 3.75NLGDD465 pKa = 5.09HH466 pKa = 6.67ILTYY470 pKa = 10.98DD471 pKa = 3.57NEE473 pKa = 4.95SNPSYY478 pKa = 10.13LTVNSIRR485 pKa = 11.84EE486 pKa = 4.13SFALQDD492 pKa = 3.68YY493 pKa = 11.14LEE495 pKa = 4.39TLNAVGNRR503 pKa = 11.84YY504 pKa = 9.51VEE506 pKa = 4.17MMYY509 pKa = 11.23SMYY512 pKa = 10.64GVQVPDD518 pKa = 3.58AVVKK522 pKa = 10.58RR523 pKa = 11.84PLYY526 pKa = 10.45LNHH529 pKa = 7.48DD530 pKa = 3.9SDD532 pKa = 3.88ILKK535 pKa = 10.5ISDD538 pKa = 3.76VEE540 pKa = 4.8SNSQTLLNNNEE551 pKa = 3.93INTPLGTLAGKK562 pKa = 10.31GIAYY566 pKa = 7.78STGSGYY572 pKa = 10.8NIQASEE578 pKa = 4.22FCILISFMRR587 pKa = 11.84ISPPSIYY594 pKa = 10.41FQGLPRR600 pKa = 11.84KK601 pKa = 9.32FMHH604 pKa = 7.02FDD606 pKa = 3.35PLDD609 pKa = 4.05FYY611 pKa = 11.01NHH613 pKa = 6.15KK614 pKa = 9.84FAHH617 pKa = 6.49LGEE620 pKa = 4.36QEE622 pKa = 4.26TFQGEE627 pKa = 4.31IYY629 pKa = 9.33FTGKK633 pKa = 10.02QDD635 pKa = 4.02DD636 pKa = 3.85DD637 pKa = 4.82SKK639 pKa = 11.71VFGYY643 pKa = 8.75MPRR646 pKa = 11.84FSEE649 pKa = 3.9YY650 pKa = 9.35RR651 pKa = 11.84GQVSRR656 pKa = 11.84SCGEE660 pKa = 4.06LNTNLHH666 pKa = 5.26YY667 pKa = 10.25WLPKK671 pKa = 10.0RR672 pKa = 11.84IFNGAPKK679 pKa = 10.22LGSNFSMQHH688 pKa = 5.83PNDD691 pKa = 4.45TIDD694 pKa = 4.92MFTNWDD700 pKa = 3.44QNYY703 pKa = 10.59DD704 pKa = 3.82SIIFQSYY711 pKa = 9.67IKK713 pKa = 9.49IDD715 pKa = 3.31KK716 pKa = 10.17SSRR719 pKa = 11.84IPVYY723 pKa = 9.67PIPSHH728 pKa = 6.34LCIYY732 pKa = 10.37NHH734 pKa = 6.86
MM1 pKa = 7.63AIEE4 pKa = 4.05NSFVYY9 pKa = 10.62SLTPVDD15 pKa = 3.31QTDD18 pKa = 3.56YY19 pKa = 11.43NKK21 pKa = 10.22FDD23 pKa = 3.8FGYY26 pKa = 10.87GKK28 pKa = 10.64YY29 pKa = 8.31FTAKK33 pKa = 9.58PGKK36 pKa = 9.68FYY38 pKa = 10.23PLHH41 pKa = 5.91WEE43 pKa = 4.22EE44 pKa = 4.88IEE46 pKa = 4.23PGANIHH52 pKa = 6.93SKK54 pKa = 9.13IQHH57 pKa = 6.04FARR60 pKa = 11.84TQPLVAPSMIDD71 pKa = 3.23MNIEE75 pKa = 3.98TFSFLVPFRR84 pKa = 11.84IIFDD88 pKa = 3.76GWQEE92 pKa = 4.11FISFGDD98 pKa = 3.56GKK100 pKa = 8.06TLKK103 pKa = 10.25NDD105 pKa = 3.74LGINSQIPYY114 pKa = 8.78ITTQQLLQYY123 pKa = 9.17FAHH126 pKa = 7.33DD127 pKa = 5.35LSCPWNLGFKK137 pKa = 10.01PDD139 pKa = 3.23NCNYY143 pKa = 9.49YY144 pKa = 8.69VTYY147 pKa = 10.47KK148 pKa = 10.77DD149 pKa = 5.65LYY151 pKa = 9.21QCCEE155 pKa = 4.04LLDD158 pKa = 3.92IPMSNLIDD166 pKa = 3.44IDD168 pKa = 3.93EE169 pKa = 4.55FQSYY173 pKa = 8.2DD174 pKa = 3.62TIKK177 pKa = 10.18TFILQTFTHH186 pKa = 7.05FSLTDD191 pKa = 3.13TGQKK195 pKa = 8.81FTIDD199 pKa = 3.35QLFTSQYY206 pKa = 9.64VQIFDD211 pKa = 4.72DD212 pKa = 4.57LNIHH216 pKa = 6.24TFPVNPQNFATLHH229 pKa = 5.86QITNSASTNLAFPKK243 pKa = 10.59YY244 pKa = 9.47SFSKK248 pKa = 10.54KK249 pKa = 10.09SIWYY253 pKa = 8.33NALNLYY259 pKa = 9.51HH260 pKa = 7.11DD261 pKa = 4.78KK262 pKa = 10.71EE263 pKa = 4.56YY264 pKa = 10.9EE265 pKa = 4.07SHH267 pKa = 7.14DD268 pKa = 3.48TQLYY272 pKa = 8.26YY273 pKa = 10.85DD274 pKa = 3.82NKK276 pKa = 8.6EE277 pKa = 4.34TYY279 pKa = 7.69DD280 pKa = 3.66TSSSSLWRR288 pKa = 11.84TRR290 pKa = 11.84LLNKK294 pKa = 10.45LEE296 pKa = 4.27TNNIRR301 pKa = 11.84SYY303 pKa = 10.18WLYY306 pKa = 11.17KK307 pKa = 10.77EE308 pKa = 5.64GDD310 pKa = 4.02TKK312 pKa = 11.01PSALWFATTPQIKK325 pKa = 8.37TLNNSFKK332 pKa = 10.59FAQSKK337 pKa = 10.69INLLPFFAKK346 pKa = 10.16QKK348 pKa = 10.63VYY350 pKa = 10.83EE351 pKa = 4.09DD352 pKa = 3.43WFRR355 pKa = 11.84CEE357 pKa = 3.97PFEE360 pKa = 4.43TEE362 pKa = 3.84YY363 pKa = 11.26SEE365 pKa = 4.01YY366 pKa = 10.5TIFSGKK372 pKa = 7.53VTEE375 pKa = 4.76LPTSEE380 pKa = 4.11FVKK383 pKa = 10.67VLHH386 pKa = 6.76LSPAHH391 pKa = 5.58YY392 pKa = 10.25KK393 pKa = 9.97RR394 pKa = 11.84DD395 pKa = 3.69YY396 pKa = 10.48FQAALPQPTALQQVFVPFSSPIDD419 pKa = 3.52LPGISNPFSDD429 pKa = 4.28NRR431 pKa = 11.84PVTLQMNSDD440 pKa = 3.15KK441 pKa = 11.13TLFAEE446 pKa = 4.55YY447 pKa = 10.23KK448 pKa = 9.7PDD450 pKa = 3.57PTRR453 pKa = 11.84STIAEE458 pKa = 3.96RR459 pKa = 11.84VEE461 pKa = 3.75NLGDD465 pKa = 5.09HH466 pKa = 6.67ILTYY470 pKa = 10.98DD471 pKa = 3.57NEE473 pKa = 4.95SNPSYY478 pKa = 10.13LTVNSIRR485 pKa = 11.84EE486 pKa = 4.13SFALQDD492 pKa = 3.68YY493 pKa = 11.14LEE495 pKa = 4.39TLNAVGNRR503 pKa = 11.84YY504 pKa = 9.51VEE506 pKa = 4.17MMYY509 pKa = 11.23SMYY512 pKa = 10.64GVQVPDD518 pKa = 3.58AVVKK522 pKa = 10.58RR523 pKa = 11.84PLYY526 pKa = 10.45LNHH529 pKa = 7.48DD530 pKa = 3.9SDD532 pKa = 3.88ILKK535 pKa = 10.5ISDD538 pKa = 3.76VEE540 pKa = 4.8SNSQTLLNNNEE551 pKa = 3.93INTPLGTLAGKK562 pKa = 10.31GIAYY566 pKa = 7.78STGSGYY572 pKa = 10.8NIQASEE578 pKa = 4.22FCILISFMRR587 pKa = 11.84ISPPSIYY594 pKa = 10.41FQGLPRR600 pKa = 11.84KK601 pKa = 9.32FMHH604 pKa = 7.02FDD606 pKa = 3.35PLDD609 pKa = 4.05FYY611 pKa = 11.01NHH613 pKa = 6.15KK614 pKa = 9.84FAHH617 pKa = 6.49LGEE620 pKa = 4.36QEE622 pKa = 4.26TFQGEE627 pKa = 4.31IYY629 pKa = 9.33FTGKK633 pKa = 10.02QDD635 pKa = 4.02DD636 pKa = 3.85DD637 pKa = 4.82SKK639 pKa = 11.71VFGYY643 pKa = 8.75MPRR646 pKa = 11.84FSEE649 pKa = 3.9YY650 pKa = 9.35RR651 pKa = 11.84GQVSRR656 pKa = 11.84SCGEE660 pKa = 4.06LNTNLHH666 pKa = 5.26YY667 pKa = 10.25WLPKK671 pKa = 10.0RR672 pKa = 11.84IFNGAPKK679 pKa = 10.22LGSNFSMQHH688 pKa = 5.83PNDD691 pKa = 4.45TIDD694 pKa = 4.92MFTNWDD700 pKa = 3.44QNYY703 pKa = 10.59DD704 pKa = 3.82SIIFQSYY711 pKa = 9.67IKK713 pKa = 9.49IDD715 pKa = 3.31KK716 pKa = 10.17SSRR719 pKa = 11.84IPVYY723 pKa = 9.67PIPSHH728 pKa = 6.34LCIYY732 pKa = 10.37NHH734 pKa = 6.86
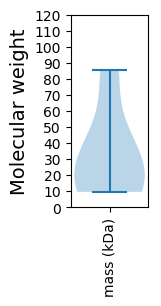
Molecular weight: 85.22 kDa
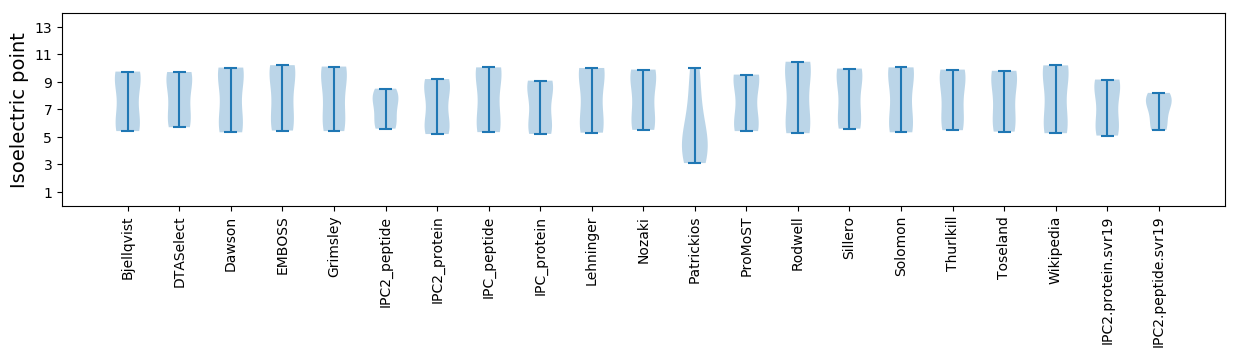
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1FVM9|A0A4V1FVM9_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_64 OX=2584787 PE=4 SV=1
MM1 pKa = 7.48KK2 pKa = 9.85CLHH5 pKa = 6.42PVGVYY10 pKa = 9.87INKK13 pKa = 9.79NYY15 pKa = 10.17KK16 pKa = 10.17DD17 pKa = 3.55LGFFQVPCGKK27 pKa = 10.4CYY29 pKa = 10.49NCKK32 pKa = 9.22QNKK35 pKa = 8.39AKK37 pKa = 10.5SVAFRR42 pKa = 11.84LEE44 pKa = 4.43LEE46 pKa = 4.23TQKK49 pKa = 11.23ASNSFFVTLTYY60 pKa = 10.02PDD62 pKa = 3.64HH63 pKa = 7.26SINKK67 pKa = 8.8INGVSCVDD75 pKa = 3.15NFITHH80 pKa = 7.56RR81 pKa = 11.84IFKK84 pKa = 10.24RR85 pKa = 11.84LRR87 pKa = 11.84SQVWNEE93 pKa = 3.67IKK95 pKa = 10.56KK96 pKa = 10.31SLHH99 pKa = 6.16KK100 pKa = 10.61LPCNRR105 pKa = 11.84FSPPSVKK112 pKa = 9.79QRR114 pKa = 11.84NRR116 pKa = 11.84QISALRR122 pKa = 11.84RR123 pKa = 11.84HH124 pKa = 6.5KK125 pKa = 11.0YY126 pKa = 10.22SLDD129 pKa = 3.92PIVRR133 pKa = 11.84SKK135 pKa = 11.4YY136 pKa = 10.58FLISEE141 pKa = 4.35YY142 pKa = 11.22GPLTSRR148 pKa = 11.84PHH150 pKa = 4.86YY151 pKa = 9.52HH152 pKa = 6.92FIWFFYY158 pKa = 10.71DD159 pKa = 3.15QVDD162 pKa = 3.63VMEE165 pKa = 4.52VEE167 pKa = 5.14KK168 pKa = 11.01KK169 pKa = 10.83LLDD172 pKa = 3.52LWDD175 pKa = 4.88GIITVDD181 pKa = 4.18LVNDD185 pKa = 3.24ARR187 pKa = 11.84INYY190 pKa = 7.88VSQYY194 pKa = 10.25CNKK197 pKa = 10.18DD198 pKa = 3.35CEE200 pKa = 4.38PQLFGQSKK208 pKa = 10.41NITRR212 pKa = 11.84CSINIGASLEE222 pKa = 3.92NSEE225 pKa = 4.28NYY227 pKa = 9.92NRR229 pKa = 11.84ILSQQCPRR237 pKa = 11.84VSWRR241 pKa = 11.84KK242 pKa = 10.03GSIKK246 pKa = 10.43LPSYY250 pKa = 9.75YY251 pKa = 9.45IRR253 pKa = 11.84RR254 pKa = 11.84AQKK257 pKa = 10.13HH258 pKa = 5.63GKK260 pKa = 8.8LQIKK264 pKa = 10.06DD265 pKa = 3.48SYY267 pKa = 11.29EE268 pKa = 4.13LYY270 pKa = 10.33TEE272 pKa = 4.38SKK274 pKa = 10.15KK275 pKa = 9.75QQQEE279 pKa = 3.36IDD281 pKa = 3.9HH282 pKa = 6.74KK283 pKa = 10.98IKK285 pKa = 10.08MYY287 pKa = 9.36CRR289 pKa = 11.84KK290 pKa = 10.11YY291 pKa = 10.64KK292 pKa = 10.28ILNKK296 pKa = 10.31HH297 pKa = 5.73EE298 pKa = 4.05FLKK301 pKa = 11.02FKK303 pKa = 10.44ISYY306 pKa = 9.86IYY308 pKa = 10.96ANEE311 pKa = 3.7KK312 pKa = 9.87NKK314 pKa = 10.31RR315 pKa = 11.84NKK317 pKa = 9.53RR318 pKa = 11.84ANII321 pKa = 3.63
MM1 pKa = 7.48KK2 pKa = 9.85CLHH5 pKa = 6.42PVGVYY10 pKa = 9.87INKK13 pKa = 9.79NYY15 pKa = 10.17KK16 pKa = 10.17DD17 pKa = 3.55LGFFQVPCGKK27 pKa = 10.4CYY29 pKa = 10.49NCKK32 pKa = 9.22QNKK35 pKa = 8.39AKK37 pKa = 10.5SVAFRR42 pKa = 11.84LEE44 pKa = 4.43LEE46 pKa = 4.23TQKK49 pKa = 11.23ASNSFFVTLTYY60 pKa = 10.02PDD62 pKa = 3.64HH63 pKa = 7.26SINKK67 pKa = 8.8INGVSCVDD75 pKa = 3.15NFITHH80 pKa = 7.56RR81 pKa = 11.84IFKK84 pKa = 10.24RR85 pKa = 11.84LRR87 pKa = 11.84SQVWNEE93 pKa = 3.67IKK95 pKa = 10.56KK96 pKa = 10.31SLHH99 pKa = 6.16KK100 pKa = 10.61LPCNRR105 pKa = 11.84FSPPSVKK112 pKa = 9.79QRR114 pKa = 11.84NRR116 pKa = 11.84QISALRR122 pKa = 11.84RR123 pKa = 11.84HH124 pKa = 6.5KK125 pKa = 11.0YY126 pKa = 10.22SLDD129 pKa = 3.92PIVRR133 pKa = 11.84SKK135 pKa = 11.4YY136 pKa = 10.58FLISEE141 pKa = 4.35YY142 pKa = 11.22GPLTSRR148 pKa = 11.84PHH150 pKa = 4.86YY151 pKa = 9.52HH152 pKa = 6.92FIWFFYY158 pKa = 10.71DD159 pKa = 3.15QVDD162 pKa = 3.63VMEE165 pKa = 4.52VEE167 pKa = 5.14KK168 pKa = 11.01KK169 pKa = 10.83LLDD172 pKa = 3.52LWDD175 pKa = 4.88GIITVDD181 pKa = 4.18LVNDD185 pKa = 3.24ARR187 pKa = 11.84INYY190 pKa = 7.88VSQYY194 pKa = 10.25CNKK197 pKa = 10.18DD198 pKa = 3.35CEE200 pKa = 4.38PQLFGQSKK208 pKa = 10.41NITRR212 pKa = 11.84CSINIGASLEE222 pKa = 3.92NSEE225 pKa = 4.28NYY227 pKa = 9.92NRR229 pKa = 11.84ILSQQCPRR237 pKa = 11.84VSWRR241 pKa = 11.84KK242 pKa = 10.03GSIKK246 pKa = 10.43LPSYY250 pKa = 9.75YY251 pKa = 9.45IRR253 pKa = 11.84RR254 pKa = 11.84AQKK257 pKa = 10.13HH258 pKa = 5.63GKK260 pKa = 8.8LQIKK264 pKa = 10.06DD265 pKa = 3.48SYY267 pKa = 11.29EE268 pKa = 4.13LYY270 pKa = 10.33TEE272 pKa = 4.38SKK274 pKa = 10.15KK275 pKa = 9.75QQQEE279 pKa = 3.36IDD281 pKa = 3.9HH282 pKa = 6.74KK283 pKa = 10.98IKK285 pKa = 10.08MYY287 pKa = 9.36CRR289 pKa = 11.84KK290 pKa = 10.11YY291 pKa = 10.64KK292 pKa = 10.28ILNKK296 pKa = 10.31HH297 pKa = 5.73EE298 pKa = 4.05FLKK301 pKa = 11.02FKK303 pKa = 10.44ISYY306 pKa = 9.86IYY308 pKa = 10.96ANEE311 pKa = 3.7KK312 pKa = 9.87NKK314 pKa = 10.31RR315 pKa = 11.84NKK317 pKa = 9.53RR318 pKa = 11.84ANII321 pKa = 3.63
Molecular weight: 38.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1645 |
79 |
734 |
274.2 |
31.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.83 ± 0.96 | 1.216 ± 0.556 |
5.775 ± 0.534 | 4.559 ± 0.341 |
6.444 ± 1.285 | 3.1 ± 0.705 |
2.553 ± 0.249 | 7.416 ± 0.432 |
7.599 ± 1.145 | 8.997 ± 0.776 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.337 ± 0.293 | 8.085 ± 0.908 |
4.316 ± 0.833 | 6.14 ± 1.189 |
3.587 ± 0.821 | 9.119 ± 0.696 |
5.775 ± 0.858 | 4.134 ± 0.608 |
0.851 ± 0.34 | 5.167 ± 0.948 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
