
Vibrio phage henriette 12B8
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; unclassified Myoviridae
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
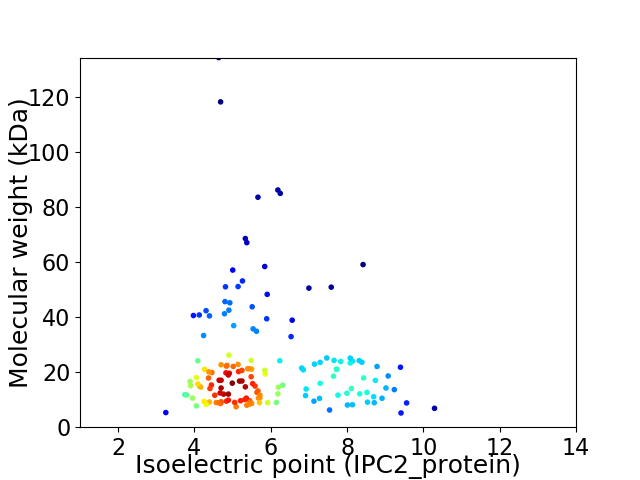
Virtual 2D-PAGE plot for 154 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M4MB99|M4MB99_9CAUD HNHc domain-containing protein OS=Vibrio phage henriette 12B8 OX=573174 GN=VPDG_00008 PE=4 SV=1
MM1 pKa = 6.85TTIKK5 pKa = 10.34LYY7 pKa = 10.79NGQTFEE13 pKa = 4.58NMSDD17 pKa = 3.49SDD19 pKa = 4.1RR20 pKa = 11.84QEE22 pKa = 3.86MATVLINRR30 pKa = 11.84EE31 pKa = 3.89FGHH34 pKa = 6.39CFSNVFSTLFAAIDD48 pKa = 3.96EE49 pKa = 4.67SHH51 pKa = 7.01VEE53 pKa = 3.91YY54 pKa = 11.08DD55 pKa = 5.7DD56 pKa = 3.77MQTLQGAYY64 pKa = 10.15DD65 pKa = 3.92YY66 pKa = 7.36TTVVTDD72 pKa = 4.83HH73 pKa = 7.07INDD76 pKa = 3.54LSLYY80 pKa = 10.75DD81 pKa = 5.01LLEE84 pKa = 4.22LVNNEE89 pKa = 4.16SLDD92 pKa = 3.63DD93 pKa = 3.87TRR95 pKa = 11.84EE96 pKa = 3.9MMHH99 pKa = 6.49TSLVKK104 pKa = 10.31DD105 pKa = 3.82LKK107 pKa = 10.77NYY109 pKa = 9.59LVEE112 pKa = 4.1YY113 pKa = 9.75VKK115 pKa = 10.36WDD117 pKa = 3.36WKK119 pKa = 10.57QSTIEE124 pKa = 4.62DD125 pKa = 3.89VFSSYY130 pKa = 11.28SGDD133 pKa = 3.41EE134 pKa = 3.96QFMHH138 pKa = 7.42DD139 pKa = 4.58EE140 pKa = 4.87DD141 pKa = 5.38IEE143 pKa = 5.64DD144 pKa = 2.89IHH146 pKa = 8.56QYY148 pKa = 8.06MDD150 pKa = 2.8RR151 pKa = 11.84VMPEE155 pKa = 3.7VVNYY159 pKa = 9.98FSDD162 pKa = 4.28IDD164 pKa = 3.86NLDD167 pKa = 3.67NFIDD171 pKa = 3.5WAEE174 pKa = 4.27TNNEE178 pKa = 3.75LSDD181 pKa = 4.27WIGTDD186 pKa = 3.29LDD188 pKa = 4.33TIRR191 pKa = 11.84AACFEE196 pKa = 5.06SISSSDD202 pKa = 4.92DD203 pKa = 3.48DD204 pKa = 4.23LQNYY208 pKa = 9.41SNNNGLDD215 pKa = 3.36AEE217 pKa = 4.22FEE219 pKa = 4.03EE220 pKa = 6.26AYY222 pKa = 9.76EE223 pKa = 4.01HH224 pKa = 6.72WIVSSWLSDD233 pKa = 3.36KK234 pKa = 11.09LPNTGDD240 pKa = 3.49VCGLTIWARR249 pKa = 11.84YY250 pKa = 7.14CTGQSICLDD259 pKa = 3.6YY260 pKa = 11.2NIQQLAFDD268 pKa = 5.24LYY270 pKa = 10.98SSEE273 pKa = 4.03VDD275 pKa = 3.01EE276 pKa = 5.23GEE278 pKa = 4.49HH279 pKa = 5.63IKK281 pKa = 10.84KK282 pKa = 10.15VDD284 pKa = 4.08AIKK287 pKa = 10.73AVIDD291 pKa = 4.0ASVKK295 pKa = 9.49PDD297 pKa = 3.13MVEE300 pKa = 3.95YY301 pKa = 9.39VSKK304 pKa = 11.14VIFDD308 pKa = 3.41NTTAYY313 pKa = 10.0QCAIDD318 pKa = 5.76ASDD321 pKa = 4.36LDD323 pKa = 3.84YY324 pKa = 11.31DD325 pKa = 4.01QRR327 pKa = 11.84KK328 pKa = 9.17ALEE331 pKa = 4.13KK332 pKa = 10.41RR333 pKa = 11.84ISSNVKK339 pKa = 9.53KK340 pKa = 10.94VNTVLEE346 pKa = 4.23SHH348 pKa = 6.35NLSLL352 pKa = 5.26
MM1 pKa = 6.85TTIKK5 pKa = 10.34LYY7 pKa = 10.79NGQTFEE13 pKa = 4.58NMSDD17 pKa = 3.49SDD19 pKa = 4.1RR20 pKa = 11.84QEE22 pKa = 3.86MATVLINRR30 pKa = 11.84EE31 pKa = 3.89FGHH34 pKa = 6.39CFSNVFSTLFAAIDD48 pKa = 3.96EE49 pKa = 4.67SHH51 pKa = 7.01VEE53 pKa = 3.91YY54 pKa = 11.08DD55 pKa = 5.7DD56 pKa = 3.77MQTLQGAYY64 pKa = 10.15DD65 pKa = 3.92YY66 pKa = 7.36TTVVTDD72 pKa = 4.83HH73 pKa = 7.07INDD76 pKa = 3.54LSLYY80 pKa = 10.75DD81 pKa = 5.01LLEE84 pKa = 4.22LVNNEE89 pKa = 4.16SLDD92 pKa = 3.63DD93 pKa = 3.87TRR95 pKa = 11.84EE96 pKa = 3.9MMHH99 pKa = 6.49TSLVKK104 pKa = 10.31DD105 pKa = 3.82LKK107 pKa = 10.77NYY109 pKa = 9.59LVEE112 pKa = 4.1YY113 pKa = 9.75VKK115 pKa = 10.36WDD117 pKa = 3.36WKK119 pKa = 10.57QSTIEE124 pKa = 4.62DD125 pKa = 3.89VFSSYY130 pKa = 11.28SGDD133 pKa = 3.41EE134 pKa = 3.96QFMHH138 pKa = 7.42DD139 pKa = 4.58EE140 pKa = 4.87DD141 pKa = 5.38IEE143 pKa = 5.64DD144 pKa = 2.89IHH146 pKa = 8.56QYY148 pKa = 8.06MDD150 pKa = 2.8RR151 pKa = 11.84VMPEE155 pKa = 3.7VVNYY159 pKa = 9.98FSDD162 pKa = 4.28IDD164 pKa = 3.86NLDD167 pKa = 3.67NFIDD171 pKa = 3.5WAEE174 pKa = 4.27TNNEE178 pKa = 3.75LSDD181 pKa = 4.27WIGTDD186 pKa = 3.29LDD188 pKa = 4.33TIRR191 pKa = 11.84AACFEE196 pKa = 5.06SISSSDD202 pKa = 4.92DD203 pKa = 3.48DD204 pKa = 4.23LQNYY208 pKa = 9.41SNNNGLDD215 pKa = 3.36AEE217 pKa = 4.22FEE219 pKa = 4.03EE220 pKa = 6.26AYY222 pKa = 9.76EE223 pKa = 4.01HH224 pKa = 6.72WIVSSWLSDD233 pKa = 3.36KK234 pKa = 11.09LPNTGDD240 pKa = 3.49VCGLTIWARR249 pKa = 11.84YY250 pKa = 7.14CTGQSICLDD259 pKa = 3.6YY260 pKa = 11.2NIQQLAFDD268 pKa = 5.24LYY270 pKa = 10.98SSEE273 pKa = 4.03VDD275 pKa = 3.01EE276 pKa = 5.23GEE278 pKa = 4.49HH279 pKa = 5.63IKK281 pKa = 10.84KK282 pKa = 10.15VDD284 pKa = 4.08AIKK287 pKa = 10.73AVIDD291 pKa = 4.0ASVKK295 pKa = 9.49PDD297 pKa = 3.13MVEE300 pKa = 3.95YY301 pKa = 9.39VSKK304 pKa = 11.14VIFDD308 pKa = 3.41NTTAYY313 pKa = 10.0QCAIDD318 pKa = 5.76ASDD321 pKa = 4.36LDD323 pKa = 3.84YY324 pKa = 11.31DD325 pKa = 4.01QRR327 pKa = 11.84KK328 pKa = 9.17ALEE331 pKa = 4.13KK332 pKa = 10.41RR333 pKa = 11.84ISSNVKK339 pKa = 9.53KK340 pKa = 10.94VNTVLEE346 pKa = 4.23SHH348 pKa = 6.35NLSLL352 pKa = 5.26
Molecular weight: 40.53 kDa
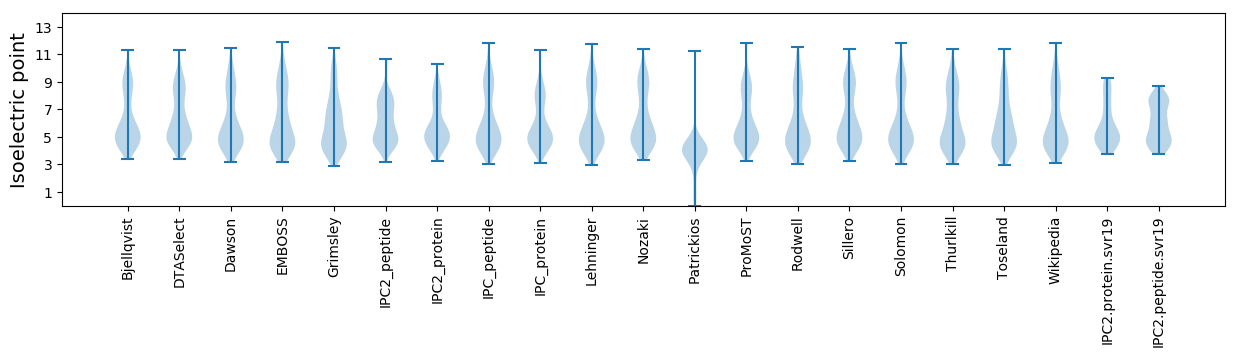
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M4MCW2|M4MCW2_9CAUD Uncharacterized protein OS=Vibrio phage henriette 12B8 OX=573174 GN=VPDG_00040 PE=4 SV=1
MM1 pKa = 7.49KK2 pKa = 10.86GNMTQEE8 pKa = 3.59YY9 pKa = 9.23KK10 pKa = 10.35IYY12 pKa = 10.54VEE14 pKa = 4.05HH15 pKa = 6.73FRR17 pKa = 11.84SKK19 pKa = 10.18RR20 pKa = 11.84CRR22 pKa = 11.84KK23 pKa = 9.31ILRR26 pKa = 11.84KK27 pKa = 8.13TYY29 pKa = 10.3RR30 pKa = 11.84FSPEE34 pKa = 3.48LAIKK38 pKa = 9.59TMHH41 pKa = 6.04QAKK44 pKa = 9.62NQLARR49 pKa = 11.84WDD51 pKa = 3.52RR52 pKa = 11.84AANKK56 pKa = 9.45LRR58 pKa = 11.84QCTRR62 pKa = 11.84DD63 pKa = 3.44YY64 pKa = 11.65DD65 pKa = 3.89VLDD68 pKa = 3.45WGVII72 pKa = 3.6
MM1 pKa = 7.49KK2 pKa = 10.86GNMTQEE8 pKa = 3.59YY9 pKa = 9.23KK10 pKa = 10.35IYY12 pKa = 10.54VEE14 pKa = 4.05HH15 pKa = 6.73FRR17 pKa = 11.84SKK19 pKa = 10.18RR20 pKa = 11.84CRR22 pKa = 11.84KK23 pKa = 9.31ILRR26 pKa = 11.84KK27 pKa = 8.13TYY29 pKa = 10.3RR30 pKa = 11.84FSPEE34 pKa = 3.48LAIKK38 pKa = 9.59TMHH41 pKa = 6.04QAKK44 pKa = 9.62NQLARR49 pKa = 11.84WDD51 pKa = 3.52RR52 pKa = 11.84AANKK56 pKa = 9.45LRR58 pKa = 11.84QCTRR62 pKa = 11.84DD63 pKa = 3.44YY64 pKa = 11.65DD65 pKa = 3.89VLDD68 pKa = 3.45WGVII72 pKa = 3.6
Molecular weight: 8.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
31838 |
42 |
1227 |
206.7 |
23.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.341 ± 0.263 | 1.684 ± 0.119 |
7.095 ± 0.147 | 6.772 ± 0.227 |
3.958 ± 0.115 | 6.637 ± 0.23 |
2.261 ± 0.103 | 6.552 ± 0.143 |
6.828 ± 0.251 | 7.513 ± 0.186 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.169 ± 0.129 | 5.613 ± 0.153 |
3.351 ± 0.146 | 3.172 ± 0.162 |
4.429 ± 0.135 | 6.238 ± 0.216 |
6.159 ± 0.251 | 6.643 ± 0.142 |
1.426 ± 0.089 | 4.159 ± 0.124 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
