
Didemnum sp. Sea Squirt associated virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.98
Get precalculated fractions of proteins
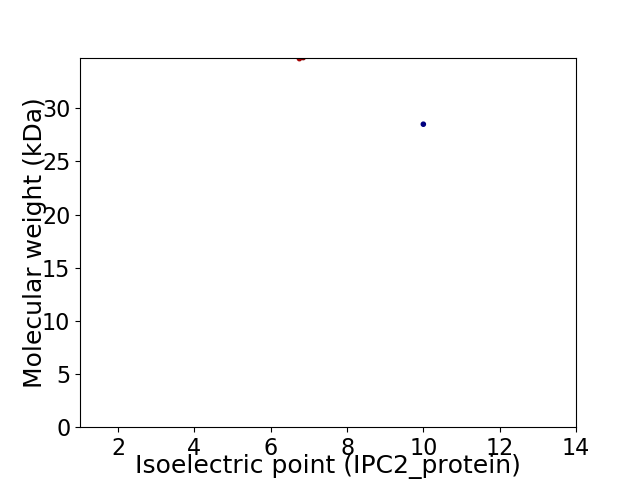
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1RL41|A0A0K1RL41_9CIRC Putative capsid protein OS=Didemnum sp. Sea Squirt associated virus OX=1692247 PE=4 SV=1
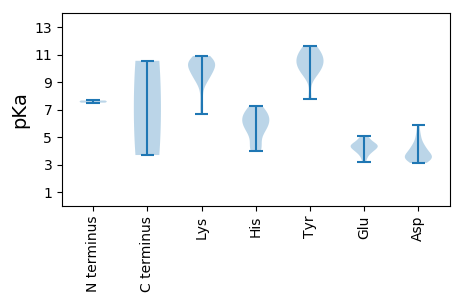
MM1 pKa = 7.66EE2 pKa = 4.36NTTTKK7 pKa = 10.69HH8 pKa = 3.99EE9 pKa = 4.49TKK11 pKa = 10.2PATNLPHH18 pKa = 7.02KK19 pKa = 10.54SWFLTLNNWTEE30 pKa = 4.05TEE32 pKa = 4.15YY33 pKa = 11.13KK34 pKa = 10.16ILQQLHH40 pKa = 5.56GVTYY44 pKa = 10.39SIIGKK49 pKa = 8.79EE50 pKa = 4.34GKK52 pKa = 7.89EE53 pKa = 4.25TTPHH57 pKa = 5.43LQGVVTFQKK66 pKa = 9.55ATRR69 pKa = 11.84IAALKK74 pKa = 10.11KK75 pKa = 9.69ILPRR79 pKa = 11.84VHH81 pKa = 6.74WEE83 pKa = 3.59IPLNLDD89 pKa = 3.56ASRR92 pKa = 11.84NYY94 pKa = 10.11CMKK97 pKa = 10.53EE98 pKa = 3.18GDD100 pKa = 4.17YY101 pKa = 11.15EE102 pKa = 4.52VTDD105 pKa = 3.58HH106 pKa = 6.59RR107 pKa = 11.84HH108 pKa = 4.36QGRR111 pKa = 11.84RR112 pKa = 11.84SDD114 pKa = 3.14IAEE117 pKa = 4.1AVGMVTDD124 pKa = 5.27GKK126 pKa = 10.51HH127 pKa = 4.25MMEE130 pKa = 4.31VAEE133 pKa = 4.52AHH135 pKa = 6.07PVTYY139 pKa = 10.44VRR141 pKa = 11.84NFRR144 pKa = 11.84GLAALQGILEE154 pKa = 4.47RR155 pKa = 11.84PRR157 pKa = 11.84TWKK160 pKa = 9.14TRR162 pKa = 11.84VHH164 pKa = 5.1VHH166 pKa = 6.45WGPTGTGKK174 pKa = 9.11TYY176 pKa = 10.53NAHH179 pKa = 6.32AQLKK183 pKa = 9.44QLFPLEE189 pKa = 4.29RR190 pKa = 11.84PYY192 pKa = 11.46VWTPAMEE199 pKa = 4.25NWWDD203 pKa = 4.14GYY205 pKa = 10.76SGQSGVILDD214 pKa = 4.01EE215 pKa = 4.88FRR217 pKa = 11.84GQLPFATLLNLLDD230 pKa = 4.57EE231 pKa = 4.96YY232 pKa = 11.63EE233 pKa = 4.46MIVPIKK239 pKa = 10.46GGSRR243 pKa = 11.84QWKK246 pKa = 7.2PHH248 pKa = 5.59VIFITSPLEE257 pKa = 3.73PRR259 pKa = 11.84SWYY262 pKa = 7.74EE263 pKa = 3.4TGKK266 pKa = 10.67AADD269 pKa = 3.64SVEE272 pKa = 3.89QLLRR276 pKa = 11.84RR277 pKa = 11.84IDD279 pKa = 3.93IIEE282 pKa = 4.55HH283 pKa = 4.11MTEE286 pKa = 3.9AYY288 pKa = 9.52KK289 pKa = 10.54ISLDD293 pKa = 5.05DD294 pKa = 3.46IEE296 pKa = 5.04QIYY299 pKa = 9.67KK300 pKa = 10.54
MM1 pKa = 7.66EE2 pKa = 4.36NTTTKK7 pKa = 10.69HH8 pKa = 3.99EE9 pKa = 4.49TKK11 pKa = 10.2PATNLPHH18 pKa = 7.02KK19 pKa = 10.54SWFLTLNNWTEE30 pKa = 4.05TEE32 pKa = 4.15YY33 pKa = 11.13KK34 pKa = 10.16ILQQLHH40 pKa = 5.56GVTYY44 pKa = 10.39SIIGKK49 pKa = 8.79EE50 pKa = 4.34GKK52 pKa = 7.89EE53 pKa = 4.25TTPHH57 pKa = 5.43LQGVVTFQKK66 pKa = 9.55ATRR69 pKa = 11.84IAALKK74 pKa = 10.11KK75 pKa = 9.69ILPRR79 pKa = 11.84VHH81 pKa = 6.74WEE83 pKa = 3.59IPLNLDD89 pKa = 3.56ASRR92 pKa = 11.84NYY94 pKa = 10.11CMKK97 pKa = 10.53EE98 pKa = 3.18GDD100 pKa = 4.17YY101 pKa = 11.15EE102 pKa = 4.52VTDD105 pKa = 3.58HH106 pKa = 6.59RR107 pKa = 11.84HH108 pKa = 4.36QGRR111 pKa = 11.84RR112 pKa = 11.84SDD114 pKa = 3.14IAEE117 pKa = 4.1AVGMVTDD124 pKa = 5.27GKK126 pKa = 10.51HH127 pKa = 4.25MMEE130 pKa = 4.31VAEE133 pKa = 4.52AHH135 pKa = 6.07PVTYY139 pKa = 10.44VRR141 pKa = 11.84NFRR144 pKa = 11.84GLAALQGILEE154 pKa = 4.47RR155 pKa = 11.84PRR157 pKa = 11.84TWKK160 pKa = 9.14TRR162 pKa = 11.84VHH164 pKa = 5.1VHH166 pKa = 6.45WGPTGTGKK174 pKa = 9.11TYY176 pKa = 10.53NAHH179 pKa = 6.32AQLKK183 pKa = 9.44QLFPLEE189 pKa = 4.29RR190 pKa = 11.84PYY192 pKa = 11.46VWTPAMEE199 pKa = 4.25NWWDD203 pKa = 4.14GYY205 pKa = 10.76SGQSGVILDD214 pKa = 4.01EE215 pKa = 4.88FRR217 pKa = 11.84GQLPFATLLNLLDD230 pKa = 4.57EE231 pKa = 4.96YY232 pKa = 11.63EE233 pKa = 4.46MIVPIKK239 pKa = 10.46GGSRR243 pKa = 11.84QWKK246 pKa = 7.2PHH248 pKa = 5.59VIFITSPLEE257 pKa = 3.73PRR259 pKa = 11.84SWYY262 pKa = 7.74EE263 pKa = 3.4TGKK266 pKa = 10.67AADD269 pKa = 3.64SVEE272 pKa = 3.89QLLRR276 pKa = 11.84RR277 pKa = 11.84IDD279 pKa = 3.93IIEE282 pKa = 4.55HH283 pKa = 4.11MTEE286 pKa = 3.9AYY288 pKa = 9.52KK289 pKa = 10.54ISLDD293 pKa = 5.05DD294 pKa = 3.46IEE296 pKa = 5.04QIYY299 pKa = 9.67KK300 pKa = 10.54
Molecular weight: 34.66 kDa
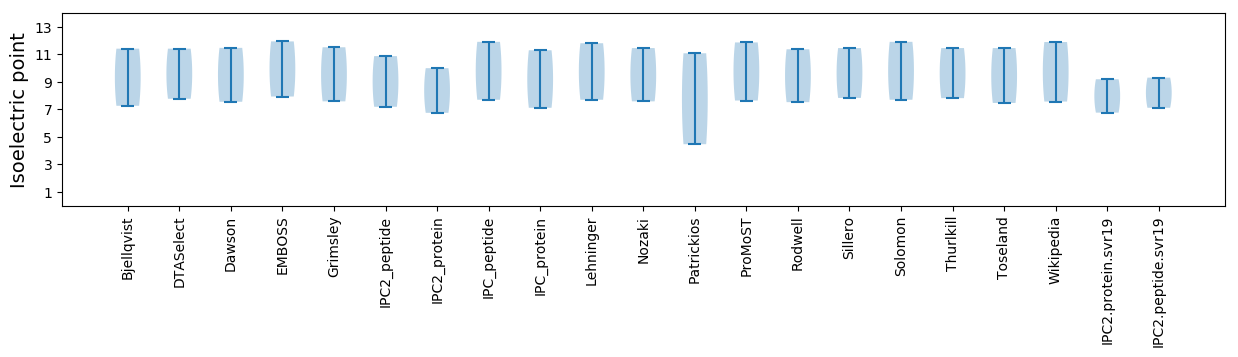
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1RL41|A0A0K1RL41_9CIRC Putative capsid protein OS=Didemnum sp. Sea Squirt associated virus OX=1692247 PE=4 SV=1
MM1 pKa = 7.5VFATPFKK8 pKa = 10.59RR9 pKa = 11.84GVKK12 pKa = 10.11RR13 pKa = 11.84LGAHH17 pKa = 6.76AARR20 pKa = 11.84SKK22 pKa = 9.99GVKK25 pKa = 9.24RR26 pKa = 11.84RR27 pKa = 11.84MFGRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84PIRR37 pKa = 11.84SRR39 pKa = 11.84FIRR42 pKa = 11.84RR43 pKa = 11.84GRR45 pKa = 11.84NLSSRR50 pKa = 11.84VRR52 pKa = 11.84TLEE55 pKa = 3.88KK56 pKa = 10.1FHH58 pKa = 5.92EE59 pKa = 4.84TKK61 pKa = 10.7FSLSNIAEE69 pKa = 4.06QSPSVGVPTSLNIGSLAVGDD89 pKa = 4.23DD90 pKa = 3.29ANTRR94 pKa = 11.84EE95 pKa = 4.3GNKK98 pKa = 9.76VSLIRR103 pKa = 11.84FDD105 pKa = 5.89LRR107 pKa = 11.84MNWVMADD114 pKa = 3.5AYY116 pKa = 10.01QAIRR120 pKa = 11.84IILVSCDD127 pKa = 3.4HH128 pKa = 6.63VSPADD133 pKa = 3.52LTLANVLDD141 pKa = 4.62ANHH144 pKa = 7.29LIARR148 pKa = 11.84LNINSSYY155 pKa = 11.02KK156 pKa = 9.61RR157 pKa = 11.84DD158 pKa = 3.55QRR160 pKa = 11.84PTPFRR165 pKa = 11.84VIWDD169 pKa = 3.42RR170 pKa = 11.84RR171 pKa = 11.84YY172 pKa = 10.55KK173 pKa = 10.24IRR175 pKa = 11.84SQTFWNTVSEE185 pKa = 4.72GVGVAPDD192 pKa = 3.6DD193 pKa = 3.76SVNIRR198 pKa = 11.84KK199 pKa = 9.43SFHH202 pKa = 6.0FPRR205 pKa = 11.84GRR207 pKa = 11.84KK208 pKa = 6.65MQFRR212 pKa = 11.84TASTDD217 pKa = 3.11NPVRR221 pKa = 11.84GGFKK225 pKa = 10.36LFYY228 pKa = 9.72MSDD231 pKa = 3.4SAAAPHH237 pKa = 5.93PTCLATTQLLFKK249 pKa = 10.9DD250 pKa = 4.28GG251 pKa = 3.69
MM1 pKa = 7.5VFATPFKK8 pKa = 10.59RR9 pKa = 11.84GVKK12 pKa = 10.11RR13 pKa = 11.84LGAHH17 pKa = 6.76AARR20 pKa = 11.84SKK22 pKa = 9.99GVKK25 pKa = 9.24RR26 pKa = 11.84RR27 pKa = 11.84MFGRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84PIRR37 pKa = 11.84SRR39 pKa = 11.84FIRR42 pKa = 11.84RR43 pKa = 11.84GRR45 pKa = 11.84NLSSRR50 pKa = 11.84VRR52 pKa = 11.84TLEE55 pKa = 3.88KK56 pKa = 10.1FHH58 pKa = 5.92EE59 pKa = 4.84TKK61 pKa = 10.7FSLSNIAEE69 pKa = 4.06QSPSVGVPTSLNIGSLAVGDD89 pKa = 4.23DD90 pKa = 3.29ANTRR94 pKa = 11.84EE95 pKa = 4.3GNKK98 pKa = 9.76VSLIRR103 pKa = 11.84FDD105 pKa = 5.89LRR107 pKa = 11.84MNWVMADD114 pKa = 3.5AYY116 pKa = 10.01QAIRR120 pKa = 11.84IILVSCDD127 pKa = 3.4HH128 pKa = 6.63VSPADD133 pKa = 3.52LTLANVLDD141 pKa = 4.62ANHH144 pKa = 7.29LIARR148 pKa = 11.84LNINSSYY155 pKa = 11.02KK156 pKa = 9.61RR157 pKa = 11.84DD158 pKa = 3.55QRR160 pKa = 11.84PTPFRR165 pKa = 11.84VIWDD169 pKa = 3.42RR170 pKa = 11.84RR171 pKa = 11.84YY172 pKa = 10.55KK173 pKa = 10.24IRR175 pKa = 11.84SQTFWNTVSEE185 pKa = 4.72GVGVAPDD192 pKa = 3.6DD193 pKa = 3.76SVNIRR198 pKa = 11.84KK199 pKa = 9.43SFHH202 pKa = 6.0FPRR205 pKa = 11.84GRR207 pKa = 11.84KK208 pKa = 6.65MQFRR212 pKa = 11.84TASTDD217 pKa = 3.11NPVRR221 pKa = 11.84GGFKK225 pKa = 10.36LFYY228 pKa = 9.72MSDD231 pKa = 3.4SAAAPHH237 pKa = 5.93PTCLATTQLLFKK249 pKa = 10.9DD250 pKa = 4.28GG251 pKa = 3.69
Molecular weight: 28.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
551 |
251 |
300 |
275.5 |
31.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.897 ± 0.69 | 0.544 ± 0.162 |
4.719 ± 0.553 | 5.263 ± 2.105 |
3.993 ± 1.276 | 6.352 ± 0.242 |
3.63 ± 0.797 | 5.808 ± 0.404 |
5.808 ± 0.404 | 7.985 ± 0.524 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.541 ± 0.097 | 4.174 ± 0.647 |
5.082 ± 0.194 | 3.448 ± 0.681 |
8.893 ± 2.481 | 5.989 ± 1.786 |
7.26 ± 1.082 | 6.352 ± 0.527 |
2.359 ± 0.749 | 2.904 ± 0.843 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
