
Aquimixticola soesokkakensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Aquimixticola
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
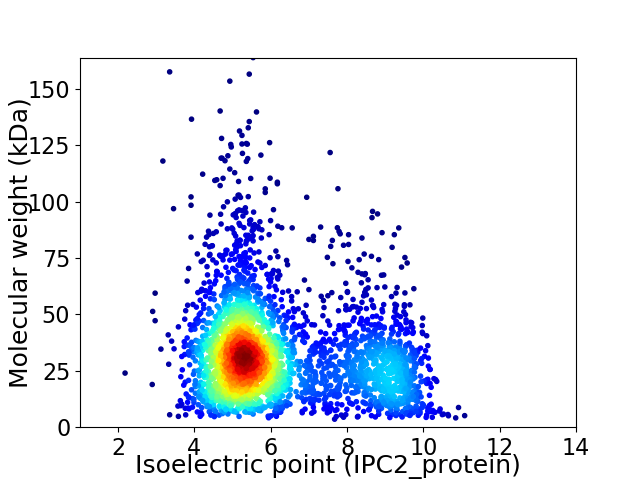
Virtual 2D-PAGE plot for 3316 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y5T5N1|A0A1Y5T5N1_9RHOB Asp/Glu/Hydantoin racemase OS=Aquimixticola soesokkakensis OX=1519096 GN=AQS8620_02469 PE=4 SV=1
MM1 pKa = 7.69KK2 pKa = 10.06ILVSSSVLALCATAAFADD20 pKa = 4.06CTEE23 pKa = 4.13VSFSDD28 pKa = 4.63VGWTDD33 pKa = 2.5ITTTTTIAKK42 pKa = 10.02QILEE46 pKa = 4.03PLGYY50 pKa = 10.17KK51 pKa = 10.05VDD53 pKa = 3.8VSVLSVPVTFASLSGGDD70 pKa = 3.29TDD72 pKa = 4.57IFLGNWMPAQSGAVTPYY89 pKa = 10.93LEE91 pKa = 5.4DD92 pKa = 3.43GTIEE96 pKa = 4.1SLHH99 pKa = 6.71EE100 pKa = 4.05NLQGTVYY107 pKa = 9.88TLAVPAYY114 pKa = 8.51TYY116 pKa = 11.04EE117 pKa = 4.55KK118 pKa = 10.34GLKK121 pKa = 10.1DD122 pKa = 3.59FADD125 pKa = 3.32IATFSDD131 pKa = 3.59ALDD134 pKa = 3.56DD135 pKa = 4.24TIYY138 pKa = 10.84GIEE141 pKa = 4.61PGNEE145 pKa = 3.73GNDD148 pKa = 3.48YY149 pKa = 10.87LISLTEE155 pKa = 4.34ADD157 pKa = 3.52THH159 pKa = 6.16GLGGFDD165 pKa = 3.28VKK167 pKa = 10.76EE168 pKa = 4.27SSEE171 pKa = 4.01QGMLSQVRR179 pKa = 11.84RR180 pKa = 11.84AYY182 pKa = 10.46DD183 pKa = 3.33KK184 pKa = 11.77NEE186 pKa = 4.86DD187 pKa = 3.47IVFLGWAPHH196 pKa = 5.62PMNVEE201 pKa = 3.86FDD203 pKa = 3.91LEE205 pKa = 4.32YY206 pKa = 10.43LTGGEE211 pKa = 4.42DD212 pKa = 3.52FFGGVGVVNTLTRR225 pKa = 11.84TGFAQEE231 pKa = 4.4CPNVGKK237 pKa = 10.59LLTNLDD243 pKa = 3.73FTVEE247 pKa = 4.08MEE249 pKa = 4.22SQIMGAILTDD259 pKa = 4.29GEE261 pKa = 4.54DD262 pKa = 3.44ADD264 pKa = 4.98DD265 pKa = 4.66AVSAYY270 pKa = 8.13ITANPGVLDD279 pKa = 3.44AWLEE283 pKa = 4.14GVTTLEE289 pKa = 4.4GDD291 pKa = 3.38AGLPAVKK298 pKa = 9.86AAYY301 pKa = 9.97GLL303 pKa = 3.86
MM1 pKa = 7.69KK2 pKa = 10.06ILVSSSVLALCATAAFADD20 pKa = 4.06CTEE23 pKa = 4.13VSFSDD28 pKa = 4.63VGWTDD33 pKa = 2.5ITTTTTIAKK42 pKa = 10.02QILEE46 pKa = 4.03PLGYY50 pKa = 10.17KK51 pKa = 10.05VDD53 pKa = 3.8VSVLSVPVTFASLSGGDD70 pKa = 3.29TDD72 pKa = 4.57IFLGNWMPAQSGAVTPYY89 pKa = 10.93LEE91 pKa = 5.4DD92 pKa = 3.43GTIEE96 pKa = 4.1SLHH99 pKa = 6.71EE100 pKa = 4.05NLQGTVYY107 pKa = 9.88TLAVPAYY114 pKa = 8.51TYY116 pKa = 11.04EE117 pKa = 4.55KK118 pKa = 10.34GLKK121 pKa = 10.1DD122 pKa = 3.59FADD125 pKa = 3.32IATFSDD131 pKa = 3.59ALDD134 pKa = 3.56DD135 pKa = 4.24TIYY138 pKa = 10.84GIEE141 pKa = 4.61PGNEE145 pKa = 3.73GNDD148 pKa = 3.48YY149 pKa = 10.87LISLTEE155 pKa = 4.34ADD157 pKa = 3.52THH159 pKa = 6.16GLGGFDD165 pKa = 3.28VKK167 pKa = 10.76EE168 pKa = 4.27SSEE171 pKa = 4.01QGMLSQVRR179 pKa = 11.84RR180 pKa = 11.84AYY182 pKa = 10.46DD183 pKa = 3.33KK184 pKa = 11.77NEE186 pKa = 4.86DD187 pKa = 3.47IVFLGWAPHH196 pKa = 5.62PMNVEE201 pKa = 3.86FDD203 pKa = 3.91LEE205 pKa = 4.32YY206 pKa = 10.43LTGGEE211 pKa = 4.42DD212 pKa = 3.52FFGGVGVVNTLTRR225 pKa = 11.84TGFAQEE231 pKa = 4.4CPNVGKK237 pKa = 10.59LLTNLDD243 pKa = 3.73FTVEE247 pKa = 4.08MEE249 pKa = 4.22SQIMGAILTDD259 pKa = 4.29GEE261 pKa = 4.54DD262 pKa = 3.44ADD264 pKa = 4.98DD265 pKa = 4.66AVSAYY270 pKa = 8.13ITANPGVLDD279 pKa = 3.44AWLEE283 pKa = 4.14GVTTLEE289 pKa = 4.4GDD291 pKa = 3.38AGLPAVKK298 pKa = 9.86AAYY301 pKa = 9.97GLL303 pKa = 3.86
Molecular weight: 32.24 kDa
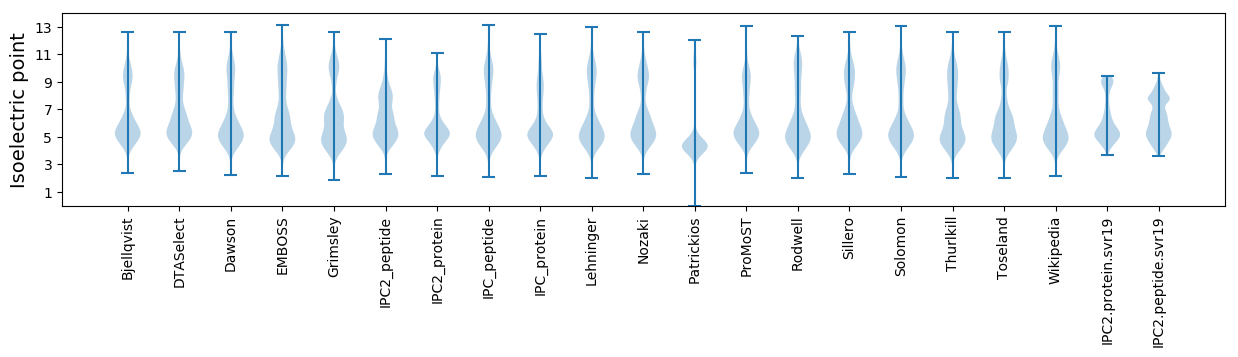
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y5SZ26|A0A1Y5SZ26_9RHOB Methylated-DNA--[protein]-cysteine S-methyltransferase OS=Aquimixticola soesokkakensis OX=1519096 GN=ada PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.81IINARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.34SLSAA44 pKa = 3.83
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.81IINARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.34SLSAA44 pKa = 3.83
Molecular weight: 5.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1038695 |
30 |
1526 |
313.2 |
33.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.89 ± 0.064 | 0.897 ± 0.011 |
6.07 ± 0.039 | 5.458 ± 0.043 |
3.786 ± 0.025 | 8.495 ± 0.04 |
1.976 ± 0.021 | 5.068 ± 0.029 |
3.282 ± 0.035 | 10.015 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.745 ± 0.022 | 2.549 ± 0.02 |
4.956 ± 0.027 | 3.422 ± 0.026 |
6.41 ± 0.047 | 5.413 ± 0.035 |
5.697 ± 0.03 | 7.382 ± 0.035 |
1.295 ± 0.016 | 2.194 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
