
Aphis glycines (Soybean aphid)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Paraneoptera; Hemiptera; Sternorrhyncha; Aphidomorpha;
Average proteome isoelectric point is 7.36
Get precalculated fractions of proteins
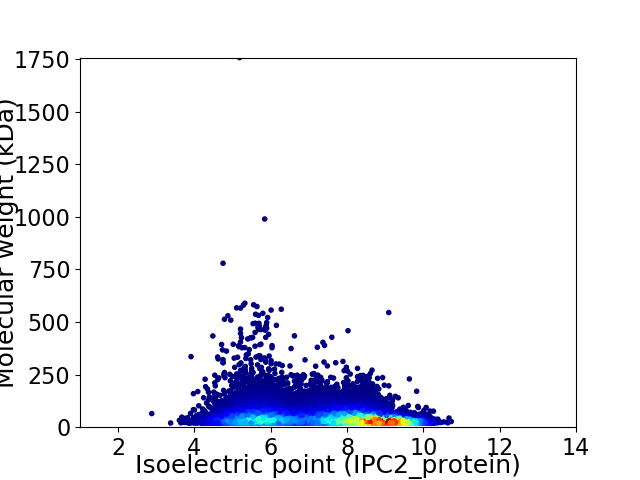
Virtual 2D-PAGE plot for 18358 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G0U8S4|A0A6G0U8S4_APHGL Uncharacterized protein OS=Aphis glycines OX=307491 GN=AGLY_000552 PE=4 SV=1
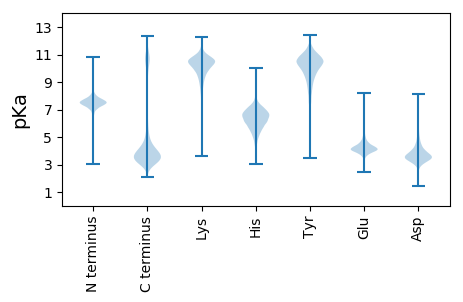
MM1 pKa = 7.52CPDD4 pKa = 3.61GTTCHH9 pKa = 6.21QNGEE13 pKa = 4.72CVRR16 pKa = 11.84STTGKK21 pKa = 9.94FSCEE25 pKa = 4.15CVVGWAGNGKK35 pKa = 8.99ICGPDD40 pKa = 3.39LDD42 pKa = 4.53LDD44 pKa = 3.93RR45 pKa = 11.84WPDD48 pKa = 3.08IDD50 pKa = 5.09LACSDD55 pKa = 4.09TRR57 pKa = 11.84CKK59 pKa = 10.44KK60 pKa = 10.29DD61 pKa = 3.08NCVRR65 pKa = 11.84VPNSGQEE72 pKa = 4.17DD73 pKa = 4.09ADD75 pKa = 3.71HH76 pKa = 7.41DD77 pKa = 5.11GIGDD81 pKa = 3.79VCDD84 pKa = 5.37DD85 pKa = 4.52DD86 pKa = 6.5ADD88 pKa = 4.05NDD90 pKa = 4.41GVPNTPDD97 pKa = 3.12NCPLVSNPDD106 pKa = 3.49QMDD109 pKa = 3.53SDD111 pKa = 4.06QDD113 pKa = 3.87GADD116 pKa = 3.81KK117 pKa = 11.19KK118 pKa = 11.36GDD120 pKa = 3.66ACDD123 pKa = 3.65NCPTIPNLDD132 pKa = 3.45QHH134 pKa = 6.76DD135 pKa = 4.08TDD137 pKa = 4.2NDD139 pKa = 5.07GIGDD143 pKa = 3.95ACDD146 pKa = 3.98ADD148 pKa = 3.68IDD150 pKa = 3.95NDD152 pKa = 4.64GILNHH157 pKa = 6.98EE158 pKa = 4.7DD159 pKa = 3.47NCPKK163 pKa = 10.36KK164 pKa = 10.77ANPDD168 pKa = 3.49QKK170 pKa = 10.88DD171 pKa = 3.19TDD173 pKa = 3.98GDD175 pKa = 4.14GLGDD179 pKa = 3.61VCDD182 pKa = 4.58NCPSMHH188 pKa = 6.76NPTQTDD194 pKa = 2.98SDD196 pKa = 3.84KK197 pKa = 11.68DD198 pKa = 4.3LIGDD202 pKa = 4.18ACDD205 pKa = 3.55SDD207 pKa = 3.39IDD209 pKa = 3.97RR210 pKa = 11.84DD211 pKa = 3.46RR212 pKa = 11.84DD213 pKa = 4.02GIQDD217 pKa = 4.22SVDD220 pKa = 3.45NCPKK224 pKa = 10.19VANSNQLDD232 pKa = 3.65TDD234 pKa = 3.87GDD236 pKa = 4.22GKK238 pKa = 11.27GDD240 pKa = 3.65EE241 pKa = 5.08CDD243 pKa = 3.85PDD245 pKa = 3.57IDD247 pKa = 5.09GDD249 pKa = 4.45GISNAQDD256 pKa = 2.92NCMLVYY262 pKa = 10.91NPDD265 pKa = 3.3QADD268 pKa = 3.37FDD270 pKa = 5.09GNGIGDD276 pKa = 3.97ICQDD280 pKa = 3.56DD281 pKa = 3.88NDD283 pKa = 3.91YY284 pKa = 11.75DD285 pKa = 3.88KK286 pKa = 11.76VPNHH290 pKa = 7.21LDD292 pKa = 3.27NCPNNSKK299 pKa = 10.26IFTTDD304 pKa = 3.09FRR306 pKa = 11.84TYY308 pKa = 8.31QTVVLDD314 pKa = 4.26PEE316 pKa = 5.01GDD318 pKa = 3.9SQIDD322 pKa = 4.05PNWVVYY328 pKa = 10.8NKK330 pKa = 10.33GAEE333 pKa = 4.33FVQTMNSDD341 pKa = 3.23PGLAVGYY348 pKa = 10.3DD349 pKa = 3.41SFGGVDD355 pKa = 3.87FEE357 pKa = 4.84GTFFVDD363 pKa = 3.1TDD365 pKa = 3.34IDD367 pKa = 3.87DD368 pKa = 5.12DD369 pKa = 4.35YY370 pKa = 11.94VGFVFSYY377 pKa = 10.52QDD379 pKa = 2.85NHH381 pKa = 6.64KK382 pKa = 10.12FYY384 pKa = 11.09TVMWKK389 pKa = 10.4KK390 pKa = 9.41GTQTYY395 pKa = 7.65WQATPFRR402 pKa = 11.84AVAEE406 pKa = 4.03PGIQIKK412 pKa = 10.05AVHH415 pKa = 6.25SEE417 pKa = 4.06TGPGQMLRR425 pKa = 11.84NALWNTEE432 pKa = 4.15STDD435 pKa = 3.19KK436 pKa = 10.48QARR439 pKa = 11.84II440 pKa = 3.86
MM1 pKa = 7.52CPDD4 pKa = 3.61GTTCHH9 pKa = 6.21QNGEE13 pKa = 4.72CVRR16 pKa = 11.84STTGKK21 pKa = 9.94FSCEE25 pKa = 4.15CVVGWAGNGKK35 pKa = 8.99ICGPDD40 pKa = 3.39LDD42 pKa = 4.53LDD44 pKa = 3.93RR45 pKa = 11.84WPDD48 pKa = 3.08IDD50 pKa = 5.09LACSDD55 pKa = 4.09TRR57 pKa = 11.84CKK59 pKa = 10.44KK60 pKa = 10.29DD61 pKa = 3.08NCVRR65 pKa = 11.84VPNSGQEE72 pKa = 4.17DD73 pKa = 4.09ADD75 pKa = 3.71HH76 pKa = 7.41DD77 pKa = 5.11GIGDD81 pKa = 3.79VCDD84 pKa = 5.37DD85 pKa = 4.52DD86 pKa = 6.5ADD88 pKa = 4.05NDD90 pKa = 4.41GVPNTPDD97 pKa = 3.12NCPLVSNPDD106 pKa = 3.49QMDD109 pKa = 3.53SDD111 pKa = 4.06QDD113 pKa = 3.87GADD116 pKa = 3.81KK117 pKa = 11.19KK118 pKa = 11.36GDD120 pKa = 3.66ACDD123 pKa = 3.65NCPTIPNLDD132 pKa = 3.45QHH134 pKa = 6.76DD135 pKa = 4.08TDD137 pKa = 4.2NDD139 pKa = 5.07GIGDD143 pKa = 3.95ACDD146 pKa = 3.98ADD148 pKa = 3.68IDD150 pKa = 3.95NDD152 pKa = 4.64GILNHH157 pKa = 6.98EE158 pKa = 4.7DD159 pKa = 3.47NCPKK163 pKa = 10.36KK164 pKa = 10.77ANPDD168 pKa = 3.49QKK170 pKa = 10.88DD171 pKa = 3.19TDD173 pKa = 3.98GDD175 pKa = 4.14GLGDD179 pKa = 3.61VCDD182 pKa = 4.58NCPSMHH188 pKa = 6.76NPTQTDD194 pKa = 2.98SDD196 pKa = 3.84KK197 pKa = 11.68DD198 pKa = 4.3LIGDD202 pKa = 4.18ACDD205 pKa = 3.55SDD207 pKa = 3.39IDD209 pKa = 3.97RR210 pKa = 11.84DD211 pKa = 3.46RR212 pKa = 11.84DD213 pKa = 4.02GIQDD217 pKa = 4.22SVDD220 pKa = 3.45NCPKK224 pKa = 10.19VANSNQLDD232 pKa = 3.65TDD234 pKa = 3.87GDD236 pKa = 4.22GKK238 pKa = 11.27GDD240 pKa = 3.65EE241 pKa = 5.08CDD243 pKa = 3.85PDD245 pKa = 3.57IDD247 pKa = 5.09GDD249 pKa = 4.45GISNAQDD256 pKa = 2.92NCMLVYY262 pKa = 10.91NPDD265 pKa = 3.3QADD268 pKa = 3.37FDD270 pKa = 5.09GNGIGDD276 pKa = 3.97ICQDD280 pKa = 3.56DD281 pKa = 3.88NDD283 pKa = 3.91YY284 pKa = 11.75DD285 pKa = 3.88KK286 pKa = 11.76VPNHH290 pKa = 7.21LDD292 pKa = 3.27NCPNNSKK299 pKa = 10.26IFTTDD304 pKa = 3.09FRR306 pKa = 11.84TYY308 pKa = 8.31QTVVLDD314 pKa = 4.26PEE316 pKa = 5.01GDD318 pKa = 3.9SQIDD322 pKa = 4.05PNWVVYY328 pKa = 10.8NKK330 pKa = 10.33GAEE333 pKa = 4.33FVQTMNSDD341 pKa = 3.23PGLAVGYY348 pKa = 10.3DD349 pKa = 3.41SFGGVDD355 pKa = 3.87FEE357 pKa = 4.84GTFFVDD363 pKa = 3.1TDD365 pKa = 3.34IDD367 pKa = 3.87DD368 pKa = 5.12DD369 pKa = 4.35YY370 pKa = 11.94VGFVFSYY377 pKa = 10.52QDD379 pKa = 2.85NHH381 pKa = 6.64KK382 pKa = 10.12FYY384 pKa = 11.09TVMWKK389 pKa = 10.4KK390 pKa = 9.41GTQTYY395 pKa = 7.65WQATPFRR402 pKa = 11.84AVAEE406 pKa = 4.03PGIQIKK412 pKa = 10.05AVHH415 pKa = 6.25SEE417 pKa = 4.06TGPGQMLRR425 pKa = 11.84NALWNTEE432 pKa = 4.15STDD435 pKa = 3.19KK436 pKa = 10.48QARR439 pKa = 11.84II440 pKa = 3.86
Molecular weight: 47.88 kDa
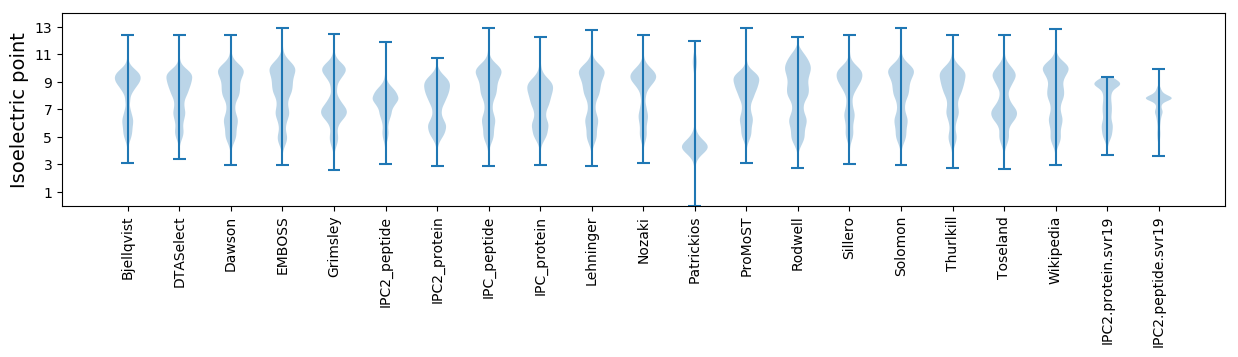
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G0U6E5|A0A6G0U6E5_APHGL AMP-binding domain-containing protein OS=Aphis glycines OX=307491 GN=AGLY_002026 PE=4 SV=1
SS1 pKa = 6.95NLPIMKK7 pKa = 10.16LNNEE11 pKa = 3.9WNIFNTATQSFRR23 pKa = 11.84RR24 pKa = 11.84ILINKK29 pKa = 8.83ILSKK33 pKa = 11.19LKK35 pKa = 9.29MLKK38 pKa = 9.51IKK40 pKa = 10.55YY41 pKa = 9.18IYY43 pKa = 10.36NIINLMCTNNCTVRR57 pKa = 11.84VLKK60 pKa = 10.84NHH62 pKa = 6.45FPTATRR68 pKa = 11.84PRR70 pKa = 11.84QTLNCLLLASQQSEE84 pKa = 4.1RR85 pKa = 11.84KK86 pKa = 9.41IIPQNDD92 pKa = 3.17KK93 pKa = 9.64LTYY96 pKa = 9.14SVYY99 pKa = 9.62MQILTGTPSVSSDD112 pKa = 4.29CPCKK116 pKa = 10.7LCTLSLIRR124 pKa = 11.84LRR126 pKa = 11.84VCTARR131 pKa = 11.84GTHH134 pKa = 5.36VDD136 pKa = 3.07SRR138 pKa = 11.84ILIRR142 pKa = 11.84FPQYY146 pKa = 9.86PGQRR150 pKa = 11.84CTFGKK155 pKa = 10.15RR156 pKa = 11.84CTNAAHH162 pKa = 6.9LHH164 pKa = 6.27HH165 pKa = 6.76NLSSEE170 pKa = 4.0CCTTVRR176 pKa = 11.84AGRR179 pKa = 11.84SNRR182 pKa = 11.84TRR184 pKa = 11.84QFLCIPLPRR193 pKa = 11.84TLEE196 pKa = 4.22DD197 pKa = 3.37CSRR200 pKa = 11.84PGICVRR206 pKa = 11.84FPIRR210 pKa = 11.84RR211 pKa = 11.84IRR213 pKa = 11.84IRR215 pKa = 11.84GSTYY219 pKa = 10.24VPIFRR224 pKa = 11.84NSTLRR229 pKa = 11.84KK230 pKa = 9.69FGWSKK235 pKa = 11.09DD236 pKa = 3.41KK237 pKa = 10.74PIRR240 pKa = 11.84SCHH243 pKa = 6.3RR244 pKa = 11.84STSFWSLQNSMACSMSCSS262 pKa = 3.43
SS1 pKa = 6.95NLPIMKK7 pKa = 10.16LNNEE11 pKa = 3.9WNIFNTATQSFRR23 pKa = 11.84RR24 pKa = 11.84ILINKK29 pKa = 8.83ILSKK33 pKa = 11.19LKK35 pKa = 9.29MLKK38 pKa = 9.51IKK40 pKa = 10.55YY41 pKa = 9.18IYY43 pKa = 10.36NIINLMCTNNCTVRR57 pKa = 11.84VLKK60 pKa = 10.84NHH62 pKa = 6.45FPTATRR68 pKa = 11.84PRR70 pKa = 11.84QTLNCLLLASQQSEE84 pKa = 4.1RR85 pKa = 11.84KK86 pKa = 9.41IIPQNDD92 pKa = 3.17KK93 pKa = 9.64LTYY96 pKa = 9.14SVYY99 pKa = 9.62MQILTGTPSVSSDD112 pKa = 4.29CPCKK116 pKa = 10.7LCTLSLIRR124 pKa = 11.84LRR126 pKa = 11.84VCTARR131 pKa = 11.84GTHH134 pKa = 5.36VDD136 pKa = 3.07SRR138 pKa = 11.84ILIRR142 pKa = 11.84FPQYY146 pKa = 9.86PGQRR150 pKa = 11.84CTFGKK155 pKa = 10.15RR156 pKa = 11.84CTNAAHH162 pKa = 6.9LHH164 pKa = 6.27HH165 pKa = 6.76NLSSEE170 pKa = 4.0CCTTVRR176 pKa = 11.84AGRR179 pKa = 11.84SNRR182 pKa = 11.84TRR184 pKa = 11.84QFLCIPLPRR193 pKa = 11.84TLEE196 pKa = 4.22DD197 pKa = 3.37CSRR200 pKa = 11.84PGICVRR206 pKa = 11.84FPIRR210 pKa = 11.84RR211 pKa = 11.84IRR213 pKa = 11.84IRR215 pKa = 11.84GSTYY219 pKa = 10.24VPIFRR224 pKa = 11.84NSTLRR229 pKa = 11.84KK230 pKa = 9.69FGWSKK235 pKa = 11.09DD236 pKa = 3.41KK237 pKa = 10.74PIRR240 pKa = 11.84SCHH243 pKa = 6.3RR244 pKa = 11.84STSFWSLQNSMACSMSCSS262 pKa = 3.43
Molecular weight: 30.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8501140 |
170 |
16471 |
463.1 |
52.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.838 ± 0.018 | 2.189 ± 0.029 |
5.278 ± 0.016 | 5.763 ± 0.02 |
4.452 ± 0.017 | 4.587 ± 0.022 |
2.487 ± 0.01 | 7.163 ± 0.02 |
7.424 ± 0.024 | 9.235 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.303 ± 0.008 | 6.474 ± 0.019 |
4.399 ± 0.027 | 3.898 ± 0.017 |
4.617 ± 0.016 | 8.229 ± 0.023 |
5.999 ± 0.017 | 5.929 ± 0.015 |
1.047 ± 0.006 | 3.687 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
