
Ilumatobacter coccineus (strain NBRC 103263 / KCTC 29153 / YM16-304)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Acidimicrobiia; Acidimicrobiales; Ilumatobacteraceae; Ilumatobacter; Ilumatobacter coccineus
Average proteome isoelectric point is 5.43
Get precalculated fractions of proteins
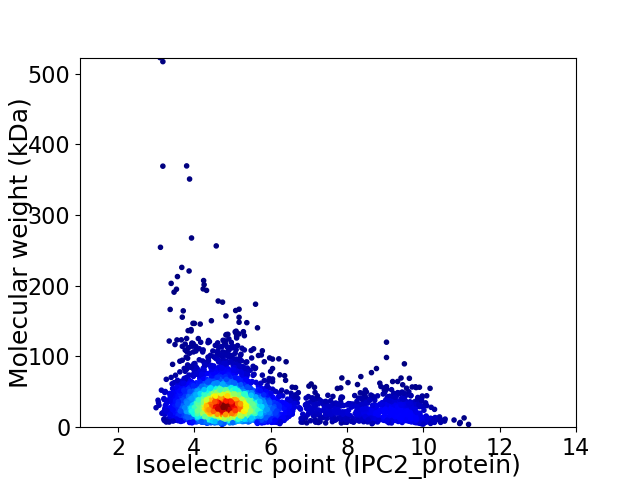
Virtual 2D-PAGE plot for 4290 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6C7E3C0|A0A6C7E3C0_ILUCY Uncharacterized protein OS=Ilumatobacter coccineus (strain NBRC 103263 / KCTC 29153 / YM16-304) OX=1313172 GN=YM304_05280 PE=4 SV=1
MM1 pKa = 7.47AAAGLAIPTAINTPIASADD20 pKa = 3.42AVGSDD25 pKa = 3.93FEE27 pKa = 4.85SFALGSVDD35 pKa = 5.24GQDD38 pKa = 3.04GWEE41 pKa = 3.79ISGPYY46 pKa = 9.55DD47 pKa = 3.68VEE49 pKa = 4.43VVDD52 pKa = 6.37ASATHH57 pKa = 6.66PSLGNRR63 pKa = 11.84SLRR66 pKa = 11.84ISNAVTSGGFGDD78 pKa = 3.81QTFSKK83 pKa = 10.25PLTEE87 pKa = 4.4EE88 pKa = 4.01AGEE91 pKa = 4.07TDD93 pKa = 3.69AEE95 pKa = 4.32NGGKK99 pKa = 8.03STGPRR104 pKa = 11.84QTSFEE109 pKa = 4.42AEE111 pKa = 3.66WSFASAAPGAEE122 pKa = 3.97QPGLSVVVSPDD133 pKa = 3.04RR134 pKa = 11.84GDD136 pKa = 3.54GGRR139 pKa = 11.84MGWLGMTDD147 pKa = 3.33TPTGLEE153 pKa = 3.89VSYY156 pKa = 11.0YY157 pKa = 10.81GYY159 pKa = 8.71DD160 pKa = 3.25TTLGGTCADD169 pKa = 3.84LDD171 pKa = 3.77NFVFTTVATGLDD183 pKa = 3.18RR184 pKa = 11.84TAVHH188 pKa = 6.03TARR191 pKa = 11.84LRR193 pKa = 11.84MGFNDD198 pKa = 4.13GVDD201 pKa = 3.39NDD203 pKa = 3.94VVEE206 pKa = 4.44VFVDD210 pKa = 4.02GTLVHH215 pKa = 6.63TGKK218 pKa = 9.9SWEE221 pKa = 4.2DD222 pKa = 3.36YY223 pKa = 10.48FRR225 pKa = 11.84NCEE228 pKa = 3.94PPEE231 pKa = 4.1PRR233 pKa = 11.84TVDD236 pKa = 3.12SLLFRR241 pKa = 11.84VSGTAAPATSGFGFLIDD258 pKa = 4.29DD259 pKa = 3.95VSLEE263 pKa = 4.24SSSPPPCTTDD273 pKa = 3.42CYY275 pKa = 11.39VDD277 pKa = 3.4AATGSDD283 pKa = 3.71VNGGTGPADD292 pKa = 3.73ALRR295 pKa = 11.84TIQTAIDD302 pKa = 3.38RR303 pKa = 11.84VDD305 pKa = 3.34PNGNVIVAAGTYY317 pKa = 7.43PVPDD321 pKa = 4.34GLDD324 pKa = 2.99VDD326 pKa = 5.03KK327 pKa = 10.99PLTLDD332 pKa = 3.52GAGSDD337 pKa = 3.82VVTIDD342 pKa = 3.66ASAHH346 pKa = 5.39ASYY349 pKa = 10.95SVAVTADD356 pKa = 3.54DD357 pKa = 3.66VTLEE361 pKa = 3.99GFTLTGNPAGTYY373 pKa = 8.69GLKK376 pKa = 10.04ISSSSPTRR384 pKa = 11.84LSNITVDD391 pKa = 3.95DD392 pKa = 4.05VVVNNSKK399 pKa = 9.11RR400 pKa = 11.84TGIDD404 pKa = 3.22LNGVDD409 pKa = 5.21GGLITNVEE417 pKa = 4.17VHH419 pKa = 6.79DD420 pKa = 4.38VPSGNGLSATDD431 pKa = 3.48SNDD434 pKa = 2.79IVFEE438 pKa = 4.07NVTTSNNAWGGLAIYY453 pKa = 9.05TYY455 pKa = 10.87GRR457 pKa = 11.84YY458 pKa = 9.62YY459 pKa = 10.86SIGSNNVTIQGTNSFGEE476 pKa = 4.08ANPVYY481 pKa = 10.86VEE483 pKa = 3.99TGNYY487 pKa = 9.18NDD489 pKa = 5.1PGNPAPVTNLTVNGFDD505 pKa = 3.33YY506 pKa = 10.82TVANDD511 pKa = 3.26DD512 pKa = 3.52HH513 pKa = 6.93RR514 pKa = 11.84AGGEE518 pKa = 4.0LFTFYY523 pKa = 11.16QNTRR527 pKa = 11.84SEE529 pKa = 4.61AIEE532 pKa = 3.9FALALTNPEE541 pKa = 3.54SSYY544 pKa = 10.66VNEE547 pKa = 4.25VATGDD552 pKa = 3.64FWVGTIGFDD561 pKa = 5.01DD562 pKa = 3.97MTIQAAVDD570 pKa = 3.38HH571 pKa = 7.04AEE573 pKa = 4.2PGDD576 pKa = 3.98TIRR579 pKa = 11.84VLPGTYY585 pKa = 9.44QGNVVVDD592 pKa = 3.75EE593 pKa = 4.22HH594 pKa = 7.86VKK596 pKa = 10.63IIGSGSGSSEE606 pKa = 3.69ASNTVLQKK614 pKa = 10.5SANEE618 pKa = 3.84ALVTIQASGQSAADD632 pKa = 3.47PLLFQDD638 pKa = 5.43LRR640 pKa = 11.84LEE642 pKa = 4.16PVGEE646 pKa = 4.0YY647 pKa = 10.71GFNVNGPAPVQFVEE661 pKa = 4.21FDD663 pKa = 3.36NVKK666 pKa = 10.8VIGTNSGAAYY676 pKa = 9.18EE677 pKa = 4.46AEE679 pKa = 4.28VGLKK683 pKa = 10.52VSTNASLSDD692 pKa = 3.69VVITDD697 pKa = 3.36SAFDD701 pKa = 3.8DD702 pKa = 5.39LVYY705 pKa = 10.46GWYY708 pKa = 9.24FAKK711 pKa = 10.68HH712 pKa = 6.1GDD714 pKa = 3.44WGPGGSNVSNVQVTDD729 pKa = 3.38TSFSNNEE736 pKa = 3.45VKK738 pKa = 10.72GIYY741 pKa = 8.78VEE743 pKa = 4.38KK744 pKa = 10.92LSDD747 pKa = 3.37TTFDD751 pKa = 3.17NVTVVNNGLDD761 pKa = 3.47TTFYY765 pKa = 8.93NAKK768 pKa = 8.6WNSGFDD774 pKa = 3.3INLKK778 pKa = 10.81GEE780 pKa = 4.02EE781 pKa = 4.48TYY783 pKa = 11.42QNITIVNSTFDD794 pKa = 3.64RR795 pKa = 11.84NGLGVEE801 pKa = 4.24EE802 pKa = 4.65GTGLTIKK809 pKa = 10.68ARR811 pKa = 11.84DD812 pKa = 3.54DD813 pKa = 3.66GGTYY817 pKa = 9.91GAHH820 pKa = 6.48PAEE823 pKa = 4.31LTGVTITGNDD833 pKa = 2.66ITGNEE838 pKa = 3.49RR839 pKa = 11.84GIRR842 pKa = 11.84FGEE845 pKa = 4.03PGKK848 pKa = 10.74NNATPTDD855 pKa = 3.98VIVSGNNITGNVVAGVINEE874 pKa = 4.36TLSQTNAIEE883 pKa = 4.22NWWGDD888 pKa = 3.04ASGPSGEE895 pKa = 4.64GPGTGDD901 pKa = 3.17SVSLNVSVCPWLDD914 pKa = 3.54GPAGTGNLVLDD925 pKa = 4.69DD926 pKa = 4.35CPGANEE932 pKa = 4.35LDD934 pKa = 3.93VVNTVDD940 pKa = 3.43WNGVTPDD947 pKa = 3.3AAQTFEE953 pKa = 4.03ICIEE957 pKa = 4.42GPSHH961 pKa = 7.05VVADD965 pKa = 4.06CQLAGSSTEE974 pKa = 3.94TLTWSDD980 pKa = 4.89LYY982 pKa = 11.23HH983 pKa = 7.02GDD985 pKa = 3.59YY986 pKa = 10.66TVTQTDD992 pKa = 4.21PGTEE996 pKa = 3.72WTVDD1000 pKa = 3.11ITGSPVTVADD1010 pKa = 4.66GVPSTPANVTNTHH1023 pKa = 6.81DD1024 pKa = 3.95PAEE1027 pKa = 4.79GEE1029 pKa = 4.54GPTPDD1034 pKa = 4.22APPTFPATPDD1044 pKa = 4.35DD1045 pKa = 3.7ISVVTEE1051 pKa = 4.56PGASSATVEE1060 pKa = 4.28YY1061 pKa = 9.33PLPEE1065 pKa = 4.62AADD1068 pKa = 4.63DD1069 pKa = 4.02DD1070 pKa = 4.88TEE1072 pKa = 4.24TLPTVVCTPASGTEE1086 pKa = 4.17FPVGDD1091 pKa = 3.62NTVTCTATDD1100 pKa = 3.55SADD1103 pKa = 3.44QTAEE1107 pKa = 3.94TTFTISVSPYY1117 pKa = 9.18SDD1119 pKa = 5.3LIAVDD1124 pKa = 3.69PARR1127 pKa = 11.84FLDD1130 pKa = 3.75TRR1132 pKa = 11.84GTGSNVTIDD1141 pKa = 4.94GIAQGEE1147 pKa = 4.71GPVPAGTSITIPVAEE1162 pKa = 4.93RR1163 pKa = 11.84GDD1165 pKa = 3.67VTADD1169 pKa = 3.02AVGAMVNIAAINPSGNGYY1187 pKa = 7.82VTLYY1191 pKa = 9.83PCTDD1195 pKa = 3.78EE1196 pKa = 4.95VPTAASLNFEE1206 pKa = 4.58AGVSISNATFVEE1218 pKa = 4.33LSDD1221 pKa = 5.45DD1222 pKa = 3.89GDD1224 pKa = 3.73VCAFASNTTGLALDD1238 pKa = 3.77VVGYY1242 pKa = 8.95MPVTSRR1248 pKa = 11.84IDD1250 pKa = 3.32FVAPARR1256 pKa = 11.84LLEE1259 pKa = 3.98TRR1261 pKa = 11.84IGEE1264 pKa = 4.34GLTTIDD1270 pKa = 4.62GRR1272 pKa = 11.84SQGDD1276 pKa = 3.7GAITAGNTIEE1286 pKa = 4.14VQVAGRR1292 pKa = 11.84AGVADD1297 pKa = 4.1DD1298 pKa = 5.14AVAAMVNVAAVNPTDD1313 pKa = 3.04TGYY1316 pKa = 8.45ITLFPCDD1323 pKa = 3.47SEE1325 pKa = 4.93QPVAASLNYY1334 pKa = 9.12TAGTNISNTTLVALSSDD1351 pKa = 3.19GVLCVFSEE1359 pKa = 4.44DD1360 pKa = 3.89ATDD1363 pKa = 3.65LTIDD1367 pKa = 3.76VIGYY1371 pKa = 8.85ASPGTDD1377 pKa = 2.53VGTLNPSRR1385 pKa = 11.84LLEE1388 pKa = 4.21TRR1390 pKa = 11.84TGTGQVTVDD1399 pKa = 5.23GIAQGDD1405 pKa = 4.11GQVDD1409 pKa = 3.4AGHH1412 pKa = 7.0FISVPIAGRR1421 pKa = 11.84SDD1423 pKa = 3.55VPDD1426 pKa = 3.94DD1427 pKa = 4.93AIAVVVNLAAINPAAVGYY1445 pKa = 7.75ATLYY1449 pKa = 9.96PCTEE1453 pKa = 4.31TPPVAASLNYY1463 pKa = 9.64RR1464 pKa = 11.84AGVNISNATLVDD1476 pKa = 4.28LSDD1479 pKa = 5.08DD1480 pKa = 3.8GDD1482 pKa = 3.77VCLFTSATAHH1492 pKa = 4.75YY1493 pKa = 8.53TIDD1496 pKa = 3.32VLSFVTATSVDD1507 pKa = 3.35AA1508 pKa = 5.22
MM1 pKa = 7.47AAAGLAIPTAINTPIASADD20 pKa = 3.42AVGSDD25 pKa = 3.93FEE27 pKa = 4.85SFALGSVDD35 pKa = 5.24GQDD38 pKa = 3.04GWEE41 pKa = 3.79ISGPYY46 pKa = 9.55DD47 pKa = 3.68VEE49 pKa = 4.43VVDD52 pKa = 6.37ASATHH57 pKa = 6.66PSLGNRR63 pKa = 11.84SLRR66 pKa = 11.84ISNAVTSGGFGDD78 pKa = 3.81QTFSKK83 pKa = 10.25PLTEE87 pKa = 4.4EE88 pKa = 4.01AGEE91 pKa = 4.07TDD93 pKa = 3.69AEE95 pKa = 4.32NGGKK99 pKa = 8.03STGPRR104 pKa = 11.84QTSFEE109 pKa = 4.42AEE111 pKa = 3.66WSFASAAPGAEE122 pKa = 3.97QPGLSVVVSPDD133 pKa = 3.04RR134 pKa = 11.84GDD136 pKa = 3.54GGRR139 pKa = 11.84MGWLGMTDD147 pKa = 3.33TPTGLEE153 pKa = 3.89VSYY156 pKa = 11.0YY157 pKa = 10.81GYY159 pKa = 8.71DD160 pKa = 3.25TTLGGTCADD169 pKa = 3.84LDD171 pKa = 3.77NFVFTTVATGLDD183 pKa = 3.18RR184 pKa = 11.84TAVHH188 pKa = 6.03TARR191 pKa = 11.84LRR193 pKa = 11.84MGFNDD198 pKa = 4.13GVDD201 pKa = 3.39NDD203 pKa = 3.94VVEE206 pKa = 4.44VFVDD210 pKa = 4.02GTLVHH215 pKa = 6.63TGKK218 pKa = 9.9SWEE221 pKa = 4.2DD222 pKa = 3.36YY223 pKa = 10.48FRR225 pKa = 11.84NCEE228 pKa = 3.94PPEE231 pKa = 4.1PRR233 pKa = 11.84TVDD236 pKa = 3.12SLLFRR241 pKa = 11.84VSGTAAPATSGFGFLIDD258 pKa = 4.29DD259 pKa = 3.95VSLEE263 pKa = 4.24SSSPPPCTTDD273 pKa = 3.42CYY275 pKa = 11.39VDD277 pKa = 3.4AATGSDD283 pKa = 3.71VNGGTGPADD292 pKa = 3.73ALRR295 pKa = 11.84TIQTAIDD302 pKa = 3.38RR303 pKa = 11.84VDD305 pKa = 3.34PNGNVIVAAGTYY317 pKa = 7.43PVPDD321 pKa = 4.34GLDD324 pKa = 2.99VDD326 pKa = 5.03KK327 pKa = 10.99PLTLDD332 pKa = 3.52GAGSDD337 pKa = 3.82VVTIDD342 pKa = 3.66ASAHH346 pKa = 5.39ASYY349 pKa = 10.95SVAVTADD356 pKa = 3.54DD357 pKa = 3.66VTLEE361 pKa = 3.99GFTLTGNPAGTYY373 pKa = 8.69GLKK376 pKa = 10.04ISSSSPTRR384 pKa = 11.84LSNITVDD391 pKa = 3.95DD392 pKa = 4.05VVVNNSKK399 pKa = 9.11RR400 pKa = 11.84TGIDD404 pKa = 3.22LNGVDD409 pKa = 5.21GGLITNVEE417 pKa = 4.17VHH419 pKa = 6.79DD420 pKa = 4.38VPSGNGLSATDD431 pKa = 3.48SNDD434 pKa = 2.79IVFEE438 pKa = 4.07NVTTSNNAWGGLAIYY453 pKa = 9.05TYY455 pKa = 10.87GRR457 pKa = 11.84YY458 pKa = 9.62YY459 pKa = 10.86SIGSNNVTIQGTNSFGEE476 pKa = 4.08ANPVYY481 pKa = 10.86VEE483 pKa = 3.99TGNYY487 pKa = 9.18NDD489 pKa = 5.1PGNPAPVTNLTVNGFDD505 pKa = 3.33YY506 pKa = 10.82TVANDD511 pKa = 3.26DD512 pKa = 3.52HH513 pKa = 6.93RR514 pKa = 11.84AGGEE518 pKa = 4.0LFTFYY523 pKa = 11.16QNTRR527 pKa = 11.84SEE529 pKa = 4.61AIEE532 pKa = 3.9FALALTNPEE541 pKa = 3.54SSYY544 pKa = 10.66VNEE547 pKa = 4.25VATGDD552 pKa = 3.64FWVGTIGFDD561 pKa = 5.01DD562 pKa = 3.97MTIQAAVDD570 pKa = 3.38HH571 pKa = 7.04AEE573 pKa = 4.2PGDD576 pKa = 3.98TIRR579 pKa = 11.84VLPGTYY585 pKa = 9.44QGNVVVDD592 pKa = 3.75EE593 pKa = 4.22HH594 pKa = 7.86VKK596 pKa = 10.63IIGSGSGSSEE606 pKa = 3.69ASNTVLQKK614 pKa = 10.5SANEE618 pKa = 3.84ALVTIQASGQSAADD632 pKa = 3.47PLLFQDD638 pKa = 5.43LRR640 pKa = 11.84LEE642 pKa = 4.16PVGEE646 pKa = 4.0YY647 pKa = 10.71GFNVNGPAPVQFVEE661 pKa = 4.21FDD663 pKa = 3.36NVKK666 pKa = 10.8VIGTNSGAAYY676 pKa = 9.18EE677 pKa = 4.46AEE679 pKa = 4.28VGLKK683 pKa = 10.52VSTNASLSDD692 pKa = 3.69VVITDD697 pKa = 3.36SAFDD701 pKa = 3.8DD702 pKa = 5.39LVYY705 pKa = 10.46GWYY708 pKa = 9.24FAKK711 pKa = 10.68HH712 pKa = 6.1GDD714 pKa = 3.44WGPGGSNVSNVQVTDD729 pKa = 3.38TSFSNNEE736 pKa = 3.45VKK738 pKa = 10.72GIYY741 pKa = 8.78VEE743 pKa = 4.38KK744 pKa = 10.92LSDD747 pKa = 3.37TTFDD751 pKa = 3.17NVTVVNNGLDD761 pKa = 3.47TTFYY765 pKa = 8.93NAKK768 pKa = 8.6WNSGFDD774 pKa = 3.3INLKK778 pKa = 10.81GEE780 pKa = 4.02EE781 pKa = 4.48TYY783 pKa = 11.42QNITIVNSTFDD794 pKa = 3.64RR795 pKa = 11.84NGLGVEE801 pKa = 4.24EE802 pKa = 4.65GTGLTIKK809 pKa = 10.68ARR811 pKa = 11.84DD812 pKa = 3.54DD813 pKa = 3.66GGTYY817 pKa = 9.91GAHH820 pKa = 6.48PAEE823 pKa = 4.31LTGVTITGNDD833 pKa = 2.66ITGNEE838 pKa = 3.49RR839 pKa = 11.84GIRR842 pKa = 11.84FGEE845 pKa = 4.03PGKK848 pKa = 10.74NNATPTDD855 pKa = 3.98VIVSGNNITGNVVAGVINEE874 pKa = 4.36TLSQTNAIEE883 pKa = 4.22NWWGDD888 pKa = 3.04ASGPSGEE895 pKa = 4.64GPGTGDD901 pKa = 3.17SVSLNVSVCPWLDD914 pKa = 3.54GPAGTGNLVLDD925 pKa = 4.69DD926 pKa = 4.35CPGANEE932 pKa = 4.35LDD934 pKa = 3.93VVNTVDD940 pKa = 3.43WNGVTPDD947 pKa = 3.3AAQTFEE953 pKa = 4.03ICIEE957 pKa = 4.42GPSHH961 pKa = 7.05VVADD965 pKa = 4.06CQLAGSSTEE974 pKa = 3.94TLTWSDD980 pKa = 4.89LYY982 pKa = 11.23HH983 pKa = 7.02GDD985 pKa = 3.59YY986 pKa = 10.66TVTQTDD992 pKa = 4.21PGTEE996 pKa = 3.72WTVDD1000 pKa = 3.11ITGSPVTVADD1010 pKa = 4.66GVPSTPANVTNTHH1023 pKa = 6.81DD1024 pKa = 3.95PAEE1027 pKa = 4.79GEE1029 pKa = 4.54GPTPDD1034 pKa = 4.22APPTFPATPDD1044 pKa = 4.35DD1045 pKa = 3.7ISVVTEE1051 pKa = 4.56PGASSATVEE1060 pKa = 4.28YY1061 pKa = 9.33PLPEE1065 pKa = 4.62AADD1068 pKa = 4.63DD1069 pKa = 4.02DD1070 pKa = 4.88TEE1072 pKa = 4.24TLPTVVCTPASGTEE1086 pKa = 4.17FPVGDD1091 pKa = 3.62NTVTCTATDD1100 pKa = 3.55SADD1103 pKa = 3.44QTAEE1107 pKa = 3.94TTFTISVSPYY1117 pKa = 9.18SDD1119 pKa = 5.3LIAVDD1124 pKa = 3.69PARR1127 pKa = 11.84FLDD1130 pKa = 3.75TRR1132 pKa = 11.84GTGSNVTIDD1141 pKa = 4.94GIAQGEE1147 pKa = 4.71GPVPAGTSITIPVAEE1162 pKa = 4.93RR1163 pKa = 11.84GDD1165 pKa = 3.67VTADD1169 pKa = 3.02AVGAMVNIAAINPSGNGYY1187 pKa = 7.82VTLYY1191 pKa = 9.83PCTDD1195 pKa = 3.78EE1196 pKa = 4.95VPTAASLNFEE1206 pKa = 4.58AGVSISNATFVEE1218 pKa = 4.33LSDD1221 pKa = 5.45DD1222 pKa = 3.89GDD1224 pKa = 3.73VCAFASNTTGLALDD1238 pKa = 3.77VVGYY1242 pKa = 8.95MPVTSRR1248 pKa = 11.84IDD1250 pKa = 3.32FVAPARR1256 pKa = 11.84LLEE1259 pKa = 3.98TRR1261 pKa = 11.84IGEE1264 pKa = 4.34GLTTIDD1270 pKa = 4.62GRR1272 pKa = 11.84SQGDD1276 pKa = 3.7GAITAGNTIEE1286 pKa = 4.14VQVAGRR1292 pKa = 11.84AGVADD1297 pKa = 4.1DD1298 pKa = 5.14AVAAMVNVAAVNPTDD1313 pKa = 3.04TGYY1316 pKa = 8.45ITLFPCDD1323 pKa = 3.47SEE1325 pKa = 4.93QPVAASLNYY1334 pKa = 9.12TAGTNISNTTLVALSSDD1351 pKa = 3.19GVLCVFSEE1359 pKa = 4.44DD1360 pKa = 3.89ATDD1363 pKa = 3.65LTIDD1367 pKa = 3.76VIGYY1371 pKa = 8.85ASPGTDD1377 pKa = 2.53VGTLNPSRR1385 pKa = 11.84LLEE1388 pKa = 4.21TRR1390 pKa = 11.84TGTGQVTVDD1399 pKa = 5.23GIAQGDD1405 pKa = 4.11GQVDD1409 pKa = 3.4AGHH1412 pKa = 7.0FISVPIAGRR1421 pKa = 11.84SDD1423 pKa = 3.55VPDD1426 pKa = 3.94DD1427 pKa = 4.93AIAVVVNLAAINPAAVGYY1445 pKa = 7.75ATLYY1449 pKa = 9.96PCTEE1453 pKa = 4.31TPPVAASLNYY1463 pKa = 9.64RR1464 pKa = 11.84AGVNISNATLVDD1476 pKa = 4.28LSDD1479 pKa = 5.08DD1480 pKa = 3.8GDD1482 pKa = 3.77VCLFTSATAHH1492 pKa = 4.75YY1493 pKa = 8.53TIDD1496 pKa = 3.32VLSFVTATSVDD1507 pKa = 3.35AA1508 pKa = 5.22
Molecular weight: 155.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6C7EAY4|A0A6C7EAY4_ILUCY UPF0434 protein YM304_28480 OS=Ilumatobacter coccineus (strain NBRC 103263 / KCTC 29153 / YM16-304) OX=1313172 GN=YM304_28480 PE=3 SV=1
MM1 pKa = 7.57GSLIKK6 pKa = 10.25KK7 pKa = 8.32RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.27RR11 pKa = 11.84MRR13 pKa = 11.84KK14 pKa = 8.96KK15 pKa = 9.73KK16 pKa = 8.93HH17 pKa = 5.59KK18 pKa = 10.34KK19 pKa = 6.44MLRR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84HH26 pKa = 4.1QRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.0
MM1 pKa = 7.57GSLIKK6 pKa = 10.25KK7 pKa = 8.32RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.27RR11 pKa = 11.84MRR13 pKa = 11.84KK14 pKa = 8.96KK15 pKa = 9.73KK16 pKa = 8.93HH17 pKa = 5.59KK18 pKa = 10.34KK19 pKa = 6.44MLRR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84HH26 pKa = 4.1QRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.0
Molecular weight: 3.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1492228 |
30 |
5213 |
347.8 |
37.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.205 ± 0.05 | 0.829 ± 0.012 |
7.756 ± 0.042 | 5.784 ± 0.038 |
3.203 ± 0.022 | 8.858 ± 0.043 |
2.136 ± 0.022 | 4.669 ± 0.024 |
1.83 ± 0.029 | 9.033 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.035 ± 0.019 | 2.322 ± 0.027 |
5.465 ± 0.03 | 2.566 ± 0.02 |
6.7 ± 0.055 | 5.982 ± 0.035 |
6.559 ± 0.066 | 8.821 ± 0.04 |
1.42 ± 0.015 | 1.827 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
