
Narcissus mosaic virus (NMV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Alphaflexiviridae; Potexvirus
Average proteome isoelectric point is 7.71
Get precalculated fractions of proteins
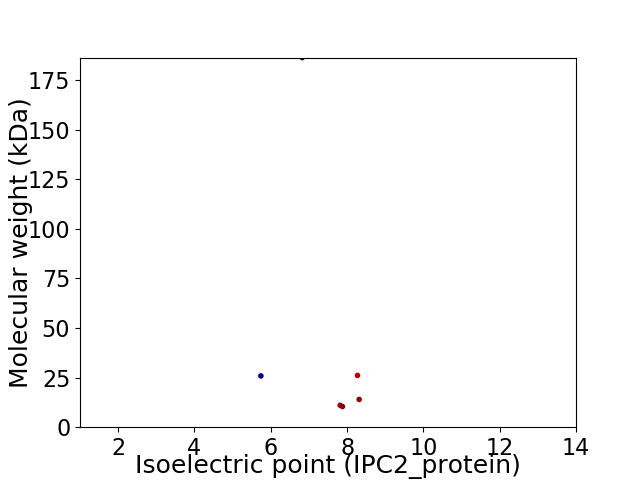
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
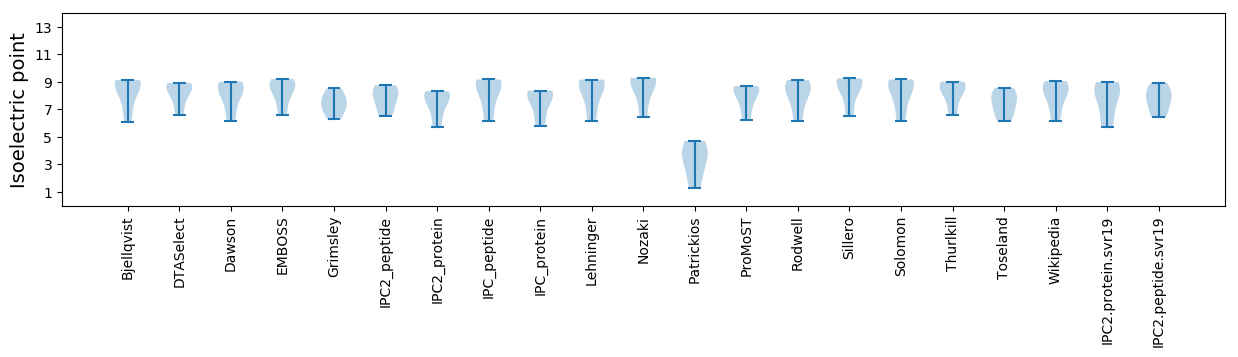
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P15097|TGB2_NMV Movement protein TGB2 OS=Narcissus mosaic virus OX=12180 GN=ORF3 PE=3 SV=1
MM1 pKa = 7.7DD2 pKa = 5.22CKK4 pKa = 11.04YY5 pKa = 10.82LLEE8 pKa = 4.91LLDD11 pKa = 4.05SYY13 pKa = 11.78SFIRR17 pKa = 11.84SSRR20 pKa = 11.84SFSSPIIIHH29 pKa = 6.32GVAGCGKK36 pKa = 8.64STIIQKK42 pKa = 9.94IALAFPEE49 pKa = 4.28LLIGSFTPALLDD61 pKa = 3.79SNSGRR66 pKa = 11.84KK67 pKa = 8.17QLAVTSDD74 pKa = 3.6PLDD77 pKa = 3.53ILDD80 pKa = 4.86EE81 pKa = 4.26YY82 pKa = 11.36LGGPNPVVRR91 pKa = 11.84LAKK94 pKa = 10.49FCDD97 pKa = 3.58PLQYY101 pKa = 10.59SCEE104 pKa = 4.13QPEE107 pKa = 4.66VPHH110 pKa = 5.6FTSLLTWRR118 pKa = 11.84FCVRR122 pKa = 11.84TTALLNGIFGCQIKK136 pKa = 10.23SRR138 pKa = 11.84RR139 pKa = 11.84EE140 pKa = 3.91DD141 pKa = 3.51LCHH144 pKa = 6.42LTHH147 pKa = 6.94EE148 pKa = 4.75NPYY151 pKa = 8.04TTDD154 pKa = 3.22PKK156 pKa = 10.93GVVVAHH162 pKa = 5.48EE163 pKa = 4.08QEE165 pKa = 4.75VINLLLQHH173 pKa = 6.81GCPVTPTQHH182 pKa = 6.34LWGLTIPVVSVYY194 pKa = 8.77ITSIASLSTVDD205 pKa = 3.81RR206 pKa = 11.84ANLFLSLTRR215 pKa = 11.84DD216 pKa = 3.26SKK218 pKa = 11.3ALHH221 pKa = 6.27IFEE224 pKa = 4.56FDD226 pKa = 3.1AWSHH230 pKa = 5.17ATCC233 pKa = 5.11
MM1 pKa = 7.7DD2 pKa = 5.22CKK4 pKa = 11.04YY5 pKa = 10.82LLEE8 pKa = 4.91LLDD11 pKa = 4.05SYY13 pKa = 11.78SFIRR17 pKa = 11.84SSRR20 pKa = 11.84SFSSPIIIHH29 pKa = 6.32GVAGCGKK36 pKa = 8.64STIIQKK42 pKa = 9.94IALAFPEE49 pKa = 4.28LLIGSFTPALLDD61 pKa = 3.79SNSGRR66 pKa = 11.84KK67 pKa = 8.17QLAVTSDD74 pKa = 3.6PLDD77 pKa = 3.53ILDD80 pKa = 4.86EE81 pKa = 4.26YY82 pKa = 11.36LGGPNPVVRR91 pKa = 11.84LAKK94 pKa = 10.49FCDD97 pKa = 3.58PLQYY101 pKa = 10.59SCEE104 pKa = 4.13QPEE107 pKa = 4.66VPHH110 pKa = 5.6FTSLLTWRR118 pKa = 11.84FCVRR122 pKa = 11.84TTALLNGIFGCQIKK136 pKa = 10.23SRR138 pKa = 11.84RR139 pKa = 11.84EE140 pKa = 3.91DD141 pKa = 3.51LCHH144 pKa = 6.42LTHH147 pKa = 6.94EE148 pKa = 4.75NPYY151 pKa = 8.04TTDD154 pKa = 3.22PKK156 pKa = 10.93GVVVAHH162 pKa = 5.48EE163 pKa = 4.08QEE165 pKa = 4.75VINLLLQHH173 pKa = 6.81GCPVTPTQHH182 pKa = 6.34LWGLTIPVVSVYY194 pKa = 8.77ITSIASLSTVDD205 pKa = 3.81RR206 pKa = 11.84ANLFLSLTRR215 pKa = 11.84DD216 pKa = 3.26SKK218 pKa = 11.3ALHH221 pKa = 6.27IFEE224 pKa = 4.56FDD226 pKa = 3.1AWSHH230 pKa = 5.17ATCC233 pKa = 5.11
Molecular weight: 25.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P15098|TGB3_NMV Movement protein TGBp3 OS=Narcissus mosaic virus OX=12180 GN=ORF4 PE=3 SV=1
MM1 pKa = 7.83PGLTPPVNYY10 pKa = 9.63EE11 pKa = 3.52QVYY14 pKa = 9.64KK15 pKa = 10.95VLAIGFLLCASIYY28 pKa = 9.71CLRR31 pKa = 11.84SNHH34 pKa = 6.1LPHH37 pKa = 7.98VGDD40 pKa = 4.89NIHH43 pKa = 6.38SLPHH47 pKa = 5.95GGNYY51 pKa = 10.23ADD53 pKa = 3.56GTKK56 pKa = 9.71RR57 pKa = 11.84VQYY60 pKa = 9.13FRR62 pKa = 11.84PHH64 pKa = 6.49SSTSTNHH71 pKa = 7.2KK72 pKa = 8.79YY73 pKa = 8.12TALCAVLTLSLLIFAQTRR91 pKa = 11.84LAAGNRR97 pKa = 11.84ITSVSICHH105 pKa = 6.31HH106 pKa = 6.74CSSQGSLSGGNHH118 pKa = 5.16GRR120 pKa = 11.84VSGHH124 pKa = 5.97SEE126 pKa = 4.06LPTTT130 pKa = 4.58
MM1 pKa = 7.83PGLTPPVNYY10 pKa = 9.63EE11 pKa = 3.52QVYY14 pKa = 9.64KK15 pKa = 10.95VLAIGFLLCASIYY28 pKa = 9.71CLRR31 pKa = 11.84SNHH34 pKa = 6.1LPHH37 pKa = 7.98VGDD40 pKa = 4.89NIHH43 pKa = 6.38SLPHH47 pKa = 5.95GGNYY51 pKa = 10.23ADD53 pKa = 3.56GTKK56 pKa = 9.71RR57 pKa = 11.84VQYY60 pKa = 9.13FRR62 pKa = 11.84PHH64 pKa = 6.49SSTSTNHH71 pKa = 7.2KK72 pKa = 8.79YY73 pKa = 8.12TALCAVLTLSLLIFAQTRR91 pKa = 11.84LAAGNRR97 pKa = 11.84ITSVSICHH105 pKa = 6.31HH106 pKa = 6.74CSSQGSLSGGNHH118 pKa = 5.16GRR120 pKa = 11.84VSGHH124 pKa = 5.97SEE126 pKa = 4.06LPTTT130 pKa = 4.58
Molecular weight: 14.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2440 |
94 |
1643 |
406.7 |
45.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.582 ± 1.086 | 2.295 ± 0.712 |
5.082 ± 0.993 | 4.959 ± 1.291 |
4.344 ± 0.532 | 4.057 ± 0.857 |
2.992 ± 0.69 | 5.369 ± 0.916 |
5.82 ± 1.274 | 10.861 ± 1.117 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.049 ± 0.528 | 3.852 ± 0.329 |
6.639 ± 0.762 | 4.139 ± 0.416 |
4.795 ± 0.155 | 8.033 ± 1.206 |
7.213 ± 0.411 | 5.615 ± 0.462 |
1.27 ± 0.186 | 3.033 ± 0.285 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
