
Alces alces faeces associated microvirus MP10 5560
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
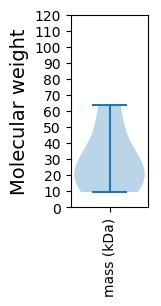
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A2Z5CI91|A0A2Z5CI91_9VIRU Minor capsid protein OS=Alces alces faeces associated microvirus MP10 5560 OX=2219133 PE=4 SV=1
MM1 pKa = 7.42NRR3 pKa = 11.84NTQNHH8 pKa = 6.26FATLPNVDD16 pKa = 3.07ISRR19 pKa = 11.84SRR21 pKa = 11.84FKK23 pKa = 11.03RR24 pKa = 11.84PFTHH28 pKa = 6.16KK29 pKa = 8.2TTFNTGDD36 pKa = 4.6IIPLMLQEE44 pKa = 4.83ILPGDD49 pKa = 4.4TVTMDD54 pKa = 3.28MGAAIRR60 pKa = 11.84MSTPIYY66 pKa = 10.2PVMDD70 pKa = 3.55NAFFDD75 pKa = 4.38YY76 pKa = 11.32YY77 pKa = 11.2FFFVPNRR84 pKa = 11.84LTWEE88 pKa = 3.57HH89 pKa = 6.3WEE91 pKa = 3.98EE92 pKa = 4.2FMGEE96 pKa = 3.99NKK98 pKa = 7.68QTAWEE103 pKa = 4.11QKK105 pKa = 8.26TEE107 pKa = 3.98YY108 pKa = 10.18EE109 pKa = 4.16IPQIKK114 pKa = 10.1APEE117 pKa = 4.51GGWAKK122 pKa = 9.5GTIADD127 pKa = 4.34HH128 pKa = 6.12MGIPTKK134 pKa = 10.74VEE136 pKa = 4.28NISVSALPFRR146 pKa = 11.84AYY148 pKa = 10.64GLIYY152 pKa = 10.65NEE154 pKa = 4.1WFRR157 pKa = 11.84DD158 pKa = 3.68QNLKK162 pKa = 10.31DD163 pKa = 3.48PCMVNTDD170 pKa = 3.95DD171 pKa = 3.87TTQTGSNSGDD181 pKa = 3.44YY182 pKa = 10.81VINTQLGALPCKK194 pKa = 9.26AAKK197 pKa = 9.53YY198 pKa = 9.96HH199 pKa = 7.56DD200 pKa = 5.05YY201 pKa = 9.17FTSALPEE208 pKa = 4.12PQKK211 pKa = 11.29GPDD214 pKa = 3.26VLLPQGLTAPVKK226 pKa = 9.7TSSEE230 pKa = 4.25YY231 pKa = 10.88AVTPEE236 pKa = 4.08EE237 pKa = 4.78VEE239 pKa = 4.33TLPSLRR245 pKa = 11.84WAGRR249 pKa = 11.84GDD251 pKa = 4.12DD252 pKa = 4.48GVTTYY257 pKa = 10.03TGSRR261 pKa = 11.84RR262 pKa = 11.84LVAYY266 pKa = 9.18GQGATDD272 pKa = 3.52YY273 pKa = 9.88TMEE276 pKa = 4.35GVTYY280 pKa = 10.2LAGVGDD286 pKa = 4.2STEE289 pKa = 4.27DD290 pKa = 3.33GQYY293 pKa = 10.9FPINLQADD301 pKa = 4.26LSMATGATINQLRR314 pKa = 11.84QAFAVQKK321 pKa = 10.36FYY323 pKa = 11.4EE324 pKa = 4.21KK325 pKa = 10.03QARR328 pKa = 11.84GGSRR332 pKa = 11.84YY333 pKa = 9.63RR334 pKa = 11.84EE335 pKa = 4.46TIRR338 pKa = 11.84QMFGVSSPDD347 pKa = 2.78ARR349 pKa = 11.84MQIPEE354 pKa = 4.09YY355 pKa = 10.55LGGHH359 pKa = 5.7RR360 pKa = 11.84QPINIDD366 pKa = 3.25QVLQTSATDD375 pKa = 3.44TTSPQGNTAAYY386 pKa = 9.74SLTNIRR392 pKa = 11.84DD393 pKa = 3.8SVFTKK398 pKa = 10.58SFVEE402 pKa = 4.46HH403 pKa = 6.94GYY405 pKa = 11.03LMCLGVVRR413 pKa = 11.84TYY415 pKa = 9.42HH416 pKa = 6.95TYY418 pKa = 9.33QQGIDD423 pKa = 3.5AHH425 pKa = 6.12WSRR428 pKa = 11.84KK429 pKa = 6.98TLFDD433 pKa = 3.98FYY435 pKa = 11.44FPVFANLGEE444 pKa = 3.9QPIYY448 pKa = 10.81NKK450 pKa = 10.2EE451 pKa = 3.59IYY453 pKa = 9.16ATGTEE458 pKa = 3.86TDD460 pKa = 3.8EE461 pKa = 4.49EE462 pKa = 4.91VFGYY466 pKa = 9.86QEE468 pKa = 3.0AWAPYY473 pKa = 8.52RR474 pKa = 11.84YY475 pKa = 10.22APARR479 pKa = 11.84VSGAFRR485 pKa = 11.84SNYY488 pKa = 8.98EE489 pKa = 3.62QTLDD493 pKa = 3.06SWHH496 pKa = 6.04YY497 pKa = 9.34ADD499 pKa = 5.47YY500 pKa = 11.38YY501 pKa = 10.7EE502 pKa = 4.49EE503 pKa = 4.24QPILSSDD510 pKa = 3.66WIDD513 pKa = 3.27EE514 pKa = 3.75TRR516 pKa = 11.84ANVNRR521 pKa = 11.84TIAVQDD527 pKa = 3.58EE528 pKa = 4.89LEE530 pKa = 4.32DD531 pKa = 3.62QFIGDD536 pKa = 4.63FYY538 pKa = 11.41FAGDD542 pKa = 3.73WVRR545 pKa = 11.84PLPIYY550 pKa = 9.75SIPGLIDD557 pKa = 2.67HH558 pKa = 7.14HH559 pKa = 6.66
MM1 pKa = 7.42NRR3 pKa = 11.84NTQNHH8 pKa = 6.26FATLPNVDD16 pKa = 3.07ISRR19 pKa = 11.84SRR21 pKa = 11.84FKK23 pKa = 11.03RR24 pKa = 11.84PFTHH28 pKa = 6.16KK29 pKa = 8.2TTFNTGDD36 pKa = 4.6IIPLMLQEE44 pKa = 4.83ILPGDD49 pKa = 4.4TVTMDD54 pKa = 3.28MGAAIRR60 pKa = 11.84MSTPIYY66 pKa = 10.2PVMDD70 pKa = 3.55NAFFDD75 pKa = 4.38YY76 pKa = 11.32YY77 pKa = 11.2FFFVPNRR84 pKa = 11.84LTWEE88 pKa = 3.57HH89 pKa = 6.3WEE91 pKa = 3.98EE92 pKa = 4.2FMGEE96 pKa = 3.99NKK98 pKa = 7.68QTAWEE103 pKa = 4.11QKK105 pKa = 8.26TEE107 pKa = 3.98YY108 pKa = 10.18EE109 pKa = 4.16IPQIKK114 pKa = 10.1APEE117 pKa = 4.51GGWAKK122 pKa = 9.5GTIADD127 pKa = 4.34HH128 pKa = 6.12MGIPTKK134 pKa = 10.74VEE136 pKa = 4.28NISVSALPFRR146 pKa = 11.84AYY148 pKa = 10.64GLIYY152 pKa = 10.65NEE154 pKa = 4.1WFRR157 pKa = 11.84DD158 pKa = 3.68QNLKK162 pKa = 10.31DD163 pKa = 3.48PCMVNTDD170 pKa = 3.95DD171 pKa = 3.87TTQTGSNSGDD181 pKa = 3.44YY182 pKa = 10.81VINTQLGALPCKK194 pKa = 9.26AAKK197 pKa = 9.53YY198 pKa = 9.96HH199 pKa = 7.56DD200 pKa = 5.05YY201 pKa = 9.17FTSALPEE208 pKa = 4.12PQKK211 pKa = 11.29GPDD214 pKa = 3.26VLLPQGLTAPVKK226 pKa = 9.7TSSEE230 pKa = 4.25YY231 pKa = 10.88AVTPEE236 pKa = 4.08EE237 pKa = 4.78VEE239 pKa = 4.33TLPSLRR245 pKa = 11.84WAGRR249 pKa = 11.84GDD251 pKa = 4.12DD252 pKa = 4.48GVTTYY257 pKa = 10.03TGSRR261 pKa = 11.84RR262 pKa = 11.84LVAYY266 pKa = 9.18GQGATDD272 pKa = 3.52YY273 pKa = 9.88TMEE276 pKa = 4.35GVTYY280 pKa = 10.2LAGVGDD286 pKa = 4.2STEE289 pKa = 4.27DD290 pKa = 3.33GQYY293 pKa = 10.9FPINLQADD301 pKa = 4.26LSMATGATINQLRR314 pKa = 11.84QAFAVQKK321 pKa = 10.36FYY323 pKa = 11.4EE324 pKa = 4.21KK325 pKa = 10.03QARR328 pKa = 11.84GGSRR332 pKa = 11.84YY333 pKa = 9.63RR334 pKa = 11.84EE335 pKa = 4.46TIRR338 pKa = 11.84QMFGVSSPDD347 pKa = 2.78ARR349 pKa = 11.84MQIPEE354 pKa = 4.09YY355 pKa = 10.55LGGHH359 pKa = 5.7RR360 pKa = 11.84QPINIDD366 pKa = 3.25QVLQTSATDD375 pKa = 3.44TTSPQGNTAAYY386 pKa = 9.74SLTNIRR392 pKa = 11.84DD393 pKa = 3.8SVFTKK398 pKa = 10.58SFVEE402 pKa = 4.46HH403 pKa = 6.94GYY405 pKa = 11.03LMCLGVVRR413 pKa = 11.84TYY415 pKa = 9.42HH416 pKa = 6.95TYY418 pKa = 9.33QQGIDD423 pKa = 3.5AHH425 pKa = 6.12WSRR428 pKa = 11.84KK429 pKa = 6.98TLFDD433 pKa = 3.98FYY435 pKa = 11.44FPVFANLGEE444 pKa = 3.9QPIYY448 pKa = 10.81NKK450 pKa = 10.2EE451 pKa = 3.59IYY453 pKa = 9.16ATGTEE458 pKa = 3.86TDD460 pKa = 3.8EE461 pKa = 4.49EE462 pKa = 4.91VFGYY466 pKa = 9.86QEE468 pKa = 3.0AWAPYY473 pKa = 8.52RR474 pKa = 11.84YY475 pKa = 10.22APARR479 pKa = 11.84VSGAFRR485 pKa = 11.84SNYY488 pKa = 8.98EE489 pKa = 3.62QTLDD493 pKa = 3.06SWHH496 pKa = 6.04YY497 pKa = 9.34ADD499 pKa = 5.47YY500 pKa = 11.38YY501 pKa = 10.7EE502 pKa = 4.49EE503 pKa = 4.24QPILSSDD510 pKa = 3.66WIDD513 pKa = 3.27EE514 pKa = 3.75TRR516 pKa = 11.84ANVNRR521 pKa = 11.84TIAVQDD527 pKa = 3.58EE528 pKa = 4.89LEE530 pKa = 4.32DD531 pKa = 3.62QFIGDD536 pKa = 4.63FYY538 pKa = 11.41FAGDD542 pKa = 3.73WVRR545 pKa = 11.84PLPIYY550 pKa = 9.75SIPGLIDD557 pKa = 2.67HH558 pKa = 7.14HH559 pKa = 6.66
Molecular weight: 63.44 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5CHZ8|A0A2Z5CHZ8_9VIRU Major capsid protein OS=Alces alces faeces associated microvirus MP10 5560 OX=2219133 PE=3 SV=1
MM1 pKa = 7.51ACYY4 pKa = 9.69HH5 pKa = 6.59PLVAYY10 pKa = 8.07EE11 pKa = 3.98HH12 pKa = 6.56TFLKK16 pKa = 10.18TEE18 pKa = 3.99NNKK21 pKa = 10.13AVIKK25 pKa = 9.26FQKK28 pKa = 9.98PEE30 pKa = 3.67QDD32 pKa = 3.74EE33 pKa = 4.82IYY35 pKa = 10.21CGYY38 pKa = 9.12WKK40 pKa = 9.17TVEE43 pKa = 4.67LPCGTCDD50 pKa = 3.59GCKK53 pKa = 10.12LEE55 pKa = 4.87HH56 pKa = 6.49AKK58 pKa = 10.29NWAIRR63 pKa = 11.84CMLEE67 pKa = 3.72AKK69 pKa = 10.1LYY71 pKa = 7.9KK72 pKa = 10.0HH73 pKa = 6.61NYY75 pKa = 9.51FITLTYY81 pKa = 10.72AEE83 pKa = 4.75EE84 pKa = 4.14NVPTIAYY91 pKa = 8.7TDD93 pKa = 3.37ISDD96 pKa = 3.58KK97 pKa = 11.44DD98 pKa = 3.39NYY100 pKa = 10.02QYY102 pKa = 11.22CEE104 pKa = 4.05EE105 pKa = 4.16LTLCKK110 pKa = 9.76RR111 pKa = 11.84DD112 pKa = 3.39FQLFMKK118 pKa = 10.47RR119 pKa = 11.84LRR121 pKa = 11.84KK122 pKa = 9.17FYY124 pKa = 10.3KK125 pKa = 10.09KK126 pKa = 10.09KK127 pKa = 10.83YY128 pKa = 8.02KK129 pKa = 9.11WDD131 pKa = 3.93NIRR134 pKa = 11.84FFACGEE140 pKa = 4.02YY141 pKa = 10.64GDD143 pKa = 3.96RR144 pKa = 11.84TGRR147 pKa = 11.84PHH149 pKa = 4.86YY150 pKa = 10.12HH151 pKa = 7.01AIIFNLPINDD161 pKa = 4.27LKK163 pKa = 11.08PYY165 pKa = 10.06FINEE169 pKa = 4.05LHH171 pKa = 5.0QQIYY175 pKa = 9.59QSEE178 pKa = 4.66TISNIWGLGLVTIGEE193 pKa = 4.39VTYY196 pKa = 10.88EE197 pKa = 4.01SAGYY201 pKa = 8.0VARR204 pKa = 11.84YY205 pKa = 6.12TLKK208 pKa = 10.64KK209 pKa = 9.03RR210 pKa = 11.84TKK212 pKa = 10.2QEE214 pKa = 4.19TIDD217 pKa = 3.57KK218 pKa = 10.33QGEE221 pKa = 4.28KK222 pKa = 10.46EE223 pKa = 3.93FTLMSRR229 pKa = 11.84KK230 pKa = 9.54PGIAQKK236 pKa = 10.92YY237 pKa = 5.86YY238 pKa = 8.72TDD240 pKa = 3.78NVEE243 pKa = 4.94KK244 pKa = 10.1IYY246 pKa = 11.14KK247 pKa = 9.04NDD249 pKa = 3.66EE250 pKa = 4.39IIISTSLDD258 pKa = 3.4YY259 pKa = 11.0VLKK262 pKa = 10.46SKK264 pKa = 10.34PPRR267 pKa = 11.84YY268 pKa = 9.09FDD270 pKa = 3.69KK271 pKa = 11.14LYY273 pKa = 10.83DD274 pKa = 3.65IDD276 pKa = 4.97NEE278 pKa = 3.78EE279 pKa = 4.13DD280 pKa = 3.85LYY282 pKa = 11.72NKK284 pKa = 9.9KK285 pKa = 8.78QEE287 pKa = 4.09RR288 pKa = 11.84RR289 pKa = 11.84KK290 pKa = 9.83KK291 pKa = 9.26AQEE294 pKa = 3.7KK295 pKa = 10.06RR296 pKa = 11.84DD297 pKa = 3.72YY298 pKa = 8.92QQKK301 pKa = 8.36HH302 pKa = 3.75TTNRR306 pKa = 11.84PDD308 pKa = 3.56EE309 pKa = 4.15LLKK312 pKa = 10.38IAEE315 pKa = 4.28RR316 pKa = 11.84KK317 pKa = 8.76KK318 pKa = 9.9QQSTKK323 pKa = 10.91ALNRR327 pKa = 11.84QLKK330 pKa = 10.37DD331 pKa = 2.96II332 pKa = 4.62
MM1 pKa = 7.51ACYY4 pKa = 9.69HH5 pKa = 6.59PLVAYY10 pKa = 8.07EE11 pKa = 3.98HH12 pKa = 6.56TFLKK16 pKa = 10.18TEE18 pKa = 3.99NNKK21 pKa = 10.13AVIKK25 pKa = 9.26FQKK28 pKa = 9.98PEE30 pKa = 3.67QDD32 pKa = 3.74EE33 pKa = 4.82IYY35 pKa = 10.21CGYY38 pKa = 9.12WKK40 pKa = 9.17TVEE43 pKa = 4.67LPCGTCDD50 pKa = 3.59GCKK53 pKa = 10.12LEE55 pKa = 4.87HH56 pKa = 6.49AKK58 pKa = 10.29NWAIRR63 pKa = 11.84CMLEE67 pKa = 3.72AKK69 pKa = 10.1LYY71 pKa = 7.9KK72 pKa = 10.0HH73 pKa = 6.61NYY75 pKa = 9.51FITLTYY81 pKa = 10.72AEE83 pKa = 4.75EE84 pKa = 4.14NVPTIAYY91 pKa = 8.7TDD93 pKa = 3.37ISDD96 pKa = 3.58KK97 pKa = 11.44DD98 pKa = 3.39NYY100 pKa = 10.02QYY102 pKa = 11.22CEE104 pKa = 4.05EE105 pKa = 4.16LTLCKK110 pKa = 9.76RR111 pKa = 11.84DD112 pKa = 3.39FQLFMKK118 pKa = 10.47RR119 pKa = 11.84LRR121 pKa = 11.84KK122 pKa = 9.17FYY124 pKa = 10.3KK125 pKa = 10.09KK126 pKa = 10.09KK127 pKa = 10.83YY128 pKa = 8.02KK129 pKa = 9.11WDD131 pKa = 3.93NIRR134 pKa = 11.84FFACGEE140 pKa = 4.02YY141 pKa = 10.64GDD143 pKa = 3.96RR144 pKa = 11.84TGRR147 pKa = 11.84PHH149 pKa = 4.86YY150 pKa = 10.12HH151 pKa = 7.01AIIFNLPINDD161 pKa = 4.27LKK163 pKa = 11.08PYY165 pKa = 10.06FINEE169 pKa = 4.05LHH171 pKa = 5.0QQIYY175 pKa = 9.59QSEE178 pKa = 4.66TISNIWGLGLVTIGEE193 pKa = 4.39VTYY196 pKa = 10.88EE197 pKa = 4.01SAGYY201 pKa = 8.0VARR204 pKa = 11.84YY205 pKa = 6.12TLKK208 pKa = 10.64KK209 pKa = 9.03RR210 pKa = 11.84TKK212 pKa = 10.2QEE214 pKa = 4.19TIDD217 pKa = 3.57KK218 pKa = 10.33QGEE221 pKa = 4.28KK222 pKa = 10.46EE223 pKa = 3.93FTLMSRR229 pKa = 11.84KK230 pKa = 9.54PGIAQKK236 pKa = 10.92YY237 pKa = 5.86YY238 pKa = 8.72TDD240 pKa = 3.78NVEE243 pKa = 4.94KK244 pKa = 10.1IYY246 pKa = 11.14KK247 pKa = 9.04NDD249 pKa = 3.66EE250 pKa = 4.39IIISTSLDD258 pKa = 3.4YY259 pKa = 11.0VLKK262 pKa = 10.46SKK264 pKa = 10.34PPRR267 pKa = 11.84YY268 pKa = 9.09FDD270 pKa = 3.69KK271 pKa = 11.14LYY273 pKa = 10.83DD274 pKa = 3.65IDD276 pKa = 4.97NEE278 pKa = 3.78EE279 pKa = 4.13DD280 pKa = 3.85LYY282 pKa = 11.72NKK284 pKa = 9.9KK285 pKa = 8.78QEE287 pKa = 4.09RR288 pKa = 11.84RR289 pKa = 11.84KK290 pKa = 9.83KK291 pKa = 9.26AQEE294 pKa = 3.7KK295 pKa = 10.06RR296 pKa = 11.84DD297 pKa = 3.72YY298 pKa = 8.92QQKK301 pKa = 8.36HH302 pKa = 3.75TTNRR306 pKa = 11.84PDD308 pKa = 3.56EE309 pKa = 4.15LLKK312 pKa = 10.38IAEE315 pKa = 4.28RR316 pKa = 11.84KK317 pKa = 8.76KK318 pKa = 9.9QQSTKK323 pKa = 10.91ALNRR327 pKa = 11.84QLKK330 pKa = 10.37DD331 pKa = 2.96II332 pKa = 4.62
Molecular weight: 39.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1531 |
84 |
559 |
255.2 |
29.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.903 ± 1.64 | 0.98 ± 0.47 |
5.356 ± 0.681 | 7.969 ± 1.121 |
4.572 ± 0.365 | 6.532 ± 0.894 |
2.09 ± 0.385 | 6.466 ± 0.731 |
7.381 ± 2.136 | 6.14 ± 0.522 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.809 ± 0.673 | 5.683 ± 0.78 |
4.115 ± 0.75 | 5.487 ± 0.838 |
3.919 ± 0.666 | 4.768 ± 0.733 |
7.577 ± 0.68 | 3.396 ± 0.727 |
1.437 ± 0.286 | 5.421 ± 0.868 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
