
Agromyces albus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Agromyces
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
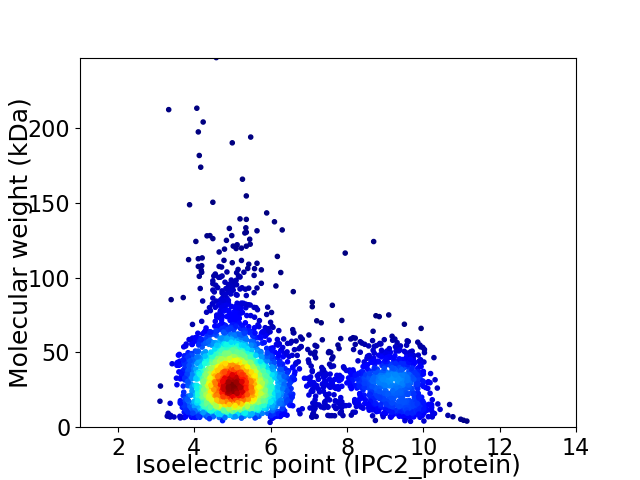
Virtual 2D-PAGE plot for 3965 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
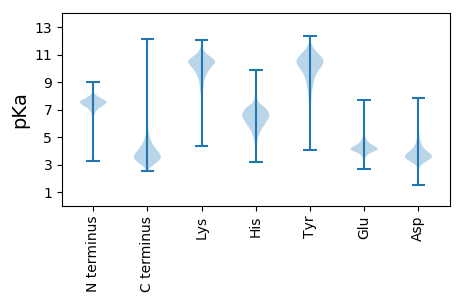
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q2KWU5|A0A4Q2KWU5_9MICO Uncharacterized protein OS=Agromyces albus OX=205332 GN=ESP51_13830 PE=4 SV=1
MM1 pKa = 7.06VSRR4 pKa = 11.84RR5 pKa = 11.84QRR7 pKa = 11.84TALLAAGALVAGLIAPVALAAPVYY31 pKa = 10.51AAVTCDD37 pKa = 3.09VGGFEE42 pKa = 5.02IDD44 pKa = 3.72GNFTAEE50 pKa = 3.88DD51 pKa = 4.12CAPGFDD57 pKa = 4.25DD58 pKa = 4.49WDD60 pKa = 3.64NVAYY64 pKa = 10.42DD65 pKa = 3.59STTQGGTYY73 pKa = 9.86SASSKK78 pKa = 10.18EE79 pKa = 4.26DD80 pKa = 3.49EE81 pKa = 5.57DD82 pKa = 4.15PAGWTSAGNTPDD94 pKa = 3.2KK95 pKa = 10.9GTFAEE100 pKa = 4.44VFSFARR106 pKa = 11.84IADD109 pKa = 3.69GDD111 pKa = 4.25YY112 pKa = 8.83FTYY115 pKa = 10.71VAWTRR120 pKa = 11.84DD121 pKa = 3.54NNTGTGGYY129 pKa = 10.21AIEE132 pKa = 4.09ITNAGEE138 pKa = 4.06RR139 pKa = 11.84VAEE142 pKa = 4.96DD143 pKa = 3.68GTPQPDD149 pKa = 3.27RR150 pKa = 11.84SLGGSVFYY158 pKa = 11.55VEE160 pKa = 4.73MQGSDD165 pKa = 3.47LPDD168 pKa = 3.86FLEE171 pKa = 4.96GCVYY175 pKa = 10.62TSVDD179 pKa = 4.31DD180 pKa = 4.3FPGVCSSDD188 pKa = 3.16EE189 pKa = 3.86TGYY192 pKa = 10.35FAEE195 pKa = 4.49ISEE198 pKa = 4.93DD199 pKa = 3.54GLFFEE204 pKa = 5.55VGLNLTEE211 pKa = 4.29LSGVAPGCPPVSEE224 pKa = 4.31TTTVYY229 pKa = 10.41VRR231 pKa = 11.84SQTGQAGTEE240 pKa = 4.08ANLKK244 pKa = 10.59GYY246 pKa = 8.13VAPLQVDD253 pKa = 4.0PPSTCGEE260 pKa = 3.87ILVDD264 pKa = 5.38KK265 pKa = 8.38EE266 pKa = 4.56TLPDD270 pKa = 3.71EE271 pKa = 4.33YY272 pKa = 11.47DD273 pKa = 3.28KK274 pKa = 11.67DD275 pKa = 4.09FEE277 pKa = 4.24FAFAGEE283 pKa = 4.37EE284 pKa = 4.22GTIDD288 pKa = 3.63FTLNDD293 pKa = 3.57ATDD296 pKa = 3.94DD297 pKa = 4.14EE298 pKa = 4.87GALWSSGPIPAGTYY312 pKa = 7.47TLEE315 pKa = 4.17EE316 pKa = 4.38LVPAGWTLDD325 pKa = 4.44DD326 pKa = 4.17ISCGTVEE333 pKa = 4.19GHH335 pKa = 5.05TTTIEE340 pKa = 3.98IAEE343 pKa = 4.44HH344 pKa = 5.8EE345 pKa = 4.59VVSCVFTNSLDD356 pKa = 3.85VVPTEE361 pKa = 3.69PAKK364 pKa = 10.37PIKK367 pKa = 8.87PTLAATGVDD376 pKa = 3.75PMSGLWLGLLLFGAGGLVFTVRR398 pKa = 11.84RR399 pKa = 11.84ARR401 pKa = 11.84ATRR404 pKa = 11.84AA405 pKa = 3.08
MM1 pKa = 7.06VSRR4 pKa = 11.84RR5 pKa = 11.84QRR7 pKa = 11.84TALLAAGALVAGLIAPVALAAPVYY31 pKa = 10.51AAVTCDD37 pKa = 3.09VGGFEE42 pKa = 5.02IDD44 pKa = 3.72GNFTAEE50 pKa = 3.88DD51 pKa = 4.12CAPGFDD57 pKa = 4.25DD58 pKa = 4.49WDD60 pKa = 3.64NVAYY64 pKa = 10.42DD65 pKa = 3.59STTQGGTYY73 pKa = 9.86SASSKK78 pKa = 10.18EE79 pKa = 4.26DD80 pKa = 3.49EE81 pKa = 5.57DD82 pKa = 4.15PAGWTSAGNTPDD94 pKa = 3.2KK95 pKa = 10.9GTFAEE100 pKa = 4.44VFSFARR106 pKa = 11.84IADD109 pKa = 3.69GDD111 pKa = 4.25YY112 pKa = 8.83FTYY115 pKa = 10.71VAWTRR120 pKa = 11.84DD121 pKa = 3.54NNTGTGGYY129 pKa = 10.21AIEE132 pKa = 4.09ITNAGEE138 pKa = 4.06RR139 pKa = 11.84VAEE142 pKa = 4.96DD143 pKa = 3.68GTPQPDD149 pKa = 3.27RR150 pKa = 11.84SLGGSVFYY158 pKa = 11.55VEE160 pKa = 4.73MQGSDD165 pKa = 3.47LPDD168 pKa = 3.86FLEE171 pKa = 4.96GCVYY175 pKa = 10.62TSVDD179 pKa = 4.31DD180 pKa = 4.3FPGVCSSDD188 pKa = 3.16EE189 pKa = 3.86TGYY192 pKa = 10.35FAEE195 pKa = 4.49ISEE198 pKa = 4.93DD199 pKa = 3.54GLFFEE204 pKa = 5.55VGLNLTEE211 pKa = 4.29LSGVAPGCPPVSEE224 pKa = 4.31TTTVYY229 pKa = 10.41VRR231 pKa = 11.84SQTGQAGTEE240 pKa = 4.08ANLKK244 pKa = 10.59GYY246 pKa = 8.13VAPLQVDD253 pKa = 4.0PPSTCGEE260 pKa = 3.87ILVDD264 pKa = 5.38KK265 pKa = 8.38EE266 pKa = 4.56TLPDD270 pKa = 3.71EE271 pKa = 4.33YY272 pKa = 11.47DD273 pKa = 3.28KK274 pKa = 11.67DD275 pKa = 4.09FEE277 pKa = 4.24FAFAGEE283 pKa = 4.37EE284 pKa = 4.22GTIDD288 pKa = 3.63FTLNDD293 pKa = 3.57ATDD296 pKa = 3.94DD297 pKa = 4.14EE298 pKa = 4.87GALWSSGPIPAGTYY312 pKa = 7.47TLEE315 pKa = 4.17EE316 pKa = 4.38LVPAGWTLDD325 pKa = 4.44DD326 pKa = 4.17ISCGTVEE333 pKa = 4.19GHH335 pKa = 5.05TTTIEE340 pKa = 3.98IAEE343 pKa = 4.44HH344 pKa = 5.8EE345 pKa = 4.59VVSCVFTNSLDD356 pKa = 3.85VVPTEE361 pKa = 3.69PAKK364 pKa = 10.37PIKK367 pKa = 8.87PTLAATGVDD376 pKa = 3.75PMSGLWLGLLLFGAGGLVFTVRR398 pKa = 11.84RR399 pKa = 11.84ARR401 pKa = 11.84ATRR404 pKa = 11.84AA405 pKa = 3.08
Molecular weight: 42.63 kDa
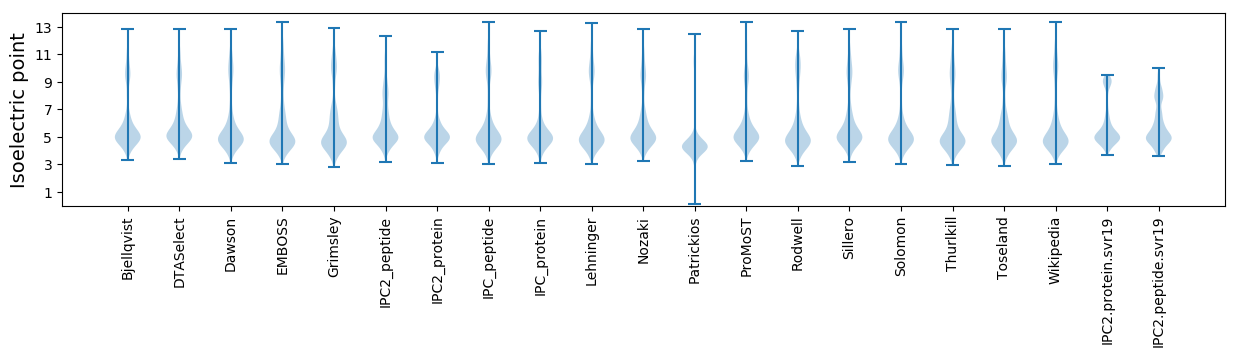
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q2L775|A0A4Q2L775_9MICO Phosphatase PAP2 family protein OS=Agromyces albus OX=205332 GN=ESP51_05305 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1288838 |
29 |
2330 |
325.1 |
34.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.563 ± 0.059 | 0.474 ± 0.009 |
6.155 ± 0.036 | 6.059 ± 0.036 |
3.289 ± 0.023 | 9.008 ± 0.035 |
2.004 ± 0.018 | 4.568 ± 0.027 |
1.761 ± 0.028 | 10.042 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.73 ± 0.013 | 1.993 ± 0.024 |
5.453 ± 0.029 | 2.563 ± 0.017 |
7.324 ± 0.044 | 5.702 ± 0.028 |
5.859 ± 0.035 | 8.888 ± 0.037 |
1.555 ± 0.016 | 2.009 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
