
Molluscum contagiosum virus subtype 1 (MOCV) (MCVI)
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Pokkesviricetes; Chitovirales; Poxviridae; Chordopoxvirinae; Molluscipoxvirus; Molluscum contagiosum virus
Average proteome isoelectric point is 7.35
Get precalculated fractions of proteins
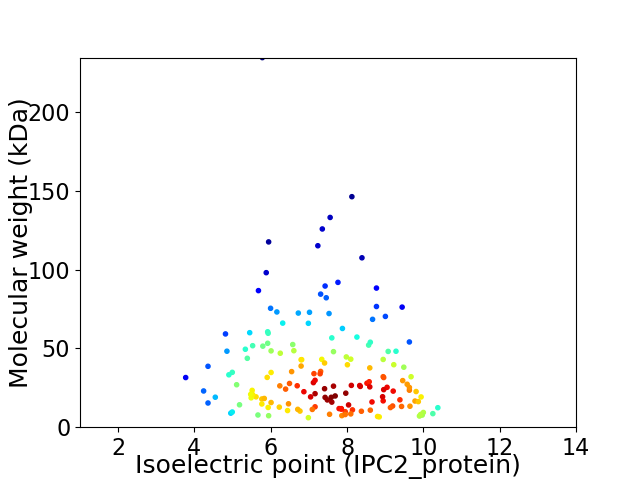
Virtual 2D-PAGE plot for 163 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q98183|Q98183_MCV1 MC012 OS=Molluscum contagiosum virus subtype 1 OX=10280 GN=MC012L PE=4 SV=1
MM1 pKa = 5.97QTCSARR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84PRR11 pKa = 11.84AAAALVVACAGALMALCAVMVSCAGDD37 pKa = 3.36VFVEE41 pKa = 4.21VSTNEE46 pKa = 3.95THH48 pKa = 7.43SLGLGSPLDD57 pKa = 4.02GDD59 pKa = 3.87GFPYY63 pKa = 10.89EE64 pKa = 4.38NFTLVLWNCTSNDD77 pKa = 3.44TAPPCGEE84 pKa = 4.91IWDD87 pKa = 3.8LWNRR91 pKa = 11.84TFPLMTLTNGTSGPEE106 pKa = 4.08VTVSGYY112 pKa = 7.54FTNQNGTCHH121 pKa = 6.44PGKK124 pKa = 10.78YY125 pKa = 9.48FGEE128 pKa = 4.49VMGSSGTSPNTLGLRR143 pKa = 11.84GLCQYY148 pKa = 10.74GNSSEE153 pKa = 4.02RR154 pKa = 11.84FPWFKK159 pKa = 10.28FVYY162 pKa = 10.49YY163 pKa = 10.55DD164 pKa = 4.57GNDD167 pKa = 3.7GNDD170 pKa = 3.46GDD172 pKa = 4.81DD173 pKa = 3.34THH175 pKa = 7.39EE176 pKa = 5.48DD177 pKa = 3.05GGEE180 pKa = 4.1YY181 pKa = 10.44YY182 pKa = 10.81DD183 pKa = 5.68GDD185 pKa = 4.23DD186 pKa = 5.19GGDD189 pKa = 3.49GGNGGNGGDD198 pKa = 4.34GGDD201 pKa = 4.4GGDD204 pKa = 5.82DD205 pKa = 4.84GDD207 pKa = 6.37DD208 pKa = 5.15DD209 pKa = 6.98DD210 pKa = 7.63DD211 pKa = 7.39DD212 pKa = 7.16DD213 pKa = 7.36GDD215 pKa = 4.5DD216 pKa = 4.02GKK218 pKa = 10.11CTFTVYY224 pKa = 11.21NLTAEE229 pKa = 4.31SLTDD233 pKa = 3.22YY234 pKa = 11.51SPIHH238 pKa = 6.29WPANTTMPPEE248 pKa = 4.01LQVRR252 pKa = 11.84LNQTGGDD259 pKa = 3.55DD260 pKa = 3.87VLNVTVPPGLFDD272 pKa = 4.86EE273 pKa = 4.71MALANATHH281 pKa = 5.42QWIGLVVNMTHH292 pKa = 6.89LCSMVV297 pKa = 2.77
MM1 pKa = 5.97QTCSARR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84PRR11 pKa = 11.84AAAALVVACAGALMALCAVMVSCAGDD37 pKa = 3.36VFVEE41 pKa = 4.21VSTNEE46 pKa = 3.95THH48 pKa = 7.43SLGLGSPLDD57 pKa = 4.02GDD59 pKa = 3.87GFPYY63 pKa = 10.89EE64 pKa = 4.38NFTLVLWNCTSNDD77 pKa = 3.44TAPPCGEE84 pKa = 4.91IWDD87 pKa = 3.8LWNRR91 pKa = 11.84TFPLMTLTNGTSGPEE106 pKa = 4.08VTVSGYY112 pKa = 7.54FTNQNGTCHH121 pKa = 6.44PGKK124 pKa = 10.78YY125 pKa = 9.48FGEE128 pKa = 4.49VMGSSGTSPNTLGLRR143 pKa = 11.84GLCQYY148 pKa = 10.74GNSSEE153 pKa = 4.02RR154 pKa = 11.84FPWFKK159 pKa = 10.28FVYY162 pKa = 10.49YY163 pKa = 10.55DD164 pKa = 4.57GNDD167 pKa = 3.7GNDD170 pKa = 3.46GDD172 pKa = 4.81DD173 pKa = 3.34THH175 pKa = 7.39EE176 pKa = 5.48DD177 pKa = 3.05GGEE180 pKa = 4.1YY181 pKa = 10.44YY182 pKa = 10.81DD183 pKa = 5.68GDD185 pKa = 4.23DD186 pKa = 5.19GGDD189 pKa = 3.49GGNGGNGGDD198 pKa = 4.34GGDD201 pKa = 4.4GGDD204 pKa = 5.82DD205 pKa = 4.84GDD207 pKa = 6.37DD208 pKa = 5.15DD209 pKa = 6.98DD210 pKa = 7.63DD211 pKa = 7.39DD212 pKa = 7.16DD213 pKa = 7.36GDD215 pKa = 4.5DD216 pKa = 4.02GKK218 pKa = 10.11CTFTVYY224 pKa = 11.21NLTAEE229 pKa = 4.31SLTDD233 pKa = 3.22YY234 pKa = 11.51SPIHH238 pKa = 6.29WPANTTMPPEE248 pKa = 4.01LQVRR252 pKa = 11.84LNQTGGDD259 pKa = 3.55DD260 pKa = 3.87VLNVTVPPGLFDD272 pKa = 4.86EE273 pKa = 4.71MALANATHH281 pKa = 5.42QWIGLVVNMTHH292 pKa = 6.89LCSMVV297 pKa = 2.77
Molecular weight: 31.5 kDa
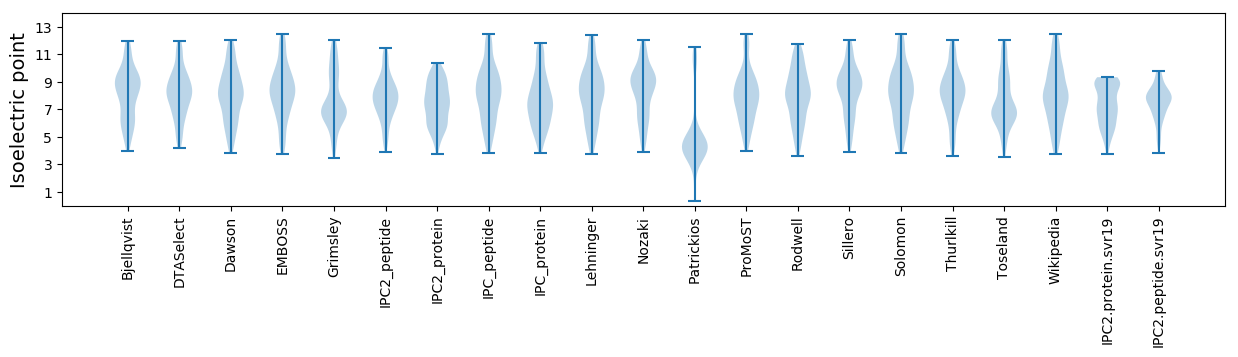
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q98291|Q98291_MCV1 MC124 OS=Molluscum contagiosum virus subtype 1 OX=10280 GN=MC124L PE=3 SV=1
MM1 pKa = 7.52SGFLAMDD8 pKa = 4.27DD9 pKa = 4.81KK10 pKa = 11.63LYY12 pKa = 10.84TDD14 pKa = 4.18LKK16 pKa = 10.3KK17 pKa = 10.89LVGAQPLYY25 pKa = 11.05LFTARR30 pKa = 11.84GDD32 pKa = 3.79FVEE35 pKa = 4.28VARR38 pKa = 11.84RR39 pKa = 11.84SEE41 pKa = 3.69FRR43 pKa = 11.84FRR45 pKa = 11.84VPAGFFAGEE54 pKa = 4.25GVALRR59 pKa = 11.84GQQFPAGSARR69 pKa = 11.84MPDD72 pKa = 3.49AAHH75 pKa = 6.56LRR77 pKa = 11.84LPRR80 pKa = 11.84LYY82 pKa = 9.75PAQQQVVAEE91 pKa = 4.2LRR93 pKa = 11.84AIAKK97 pKa = 9.8ACARR101 pKa = 11.84EE102 pKa = 3.88QRR104 pKa = 11.84PLYY107 pKa = 8.95VTLHH111 pKa = 6.28LACGFGKK118 pKa = 8.59TVTASYY124 pKa = 11.43LLGTHH129 pKa = 6.23RR130 pKa = 11.84RR131 pKa = 11.84RR132 pKa = 11.84AVVCVPTKK140 pKa = 10.3MLLAQWGAVLAALRR154 pKa = 11.84VSVYY158 pKa = 10.4VSGEE162 pKa = 4.04GVRR165 pKa = 11.84KK166 pKa = 9.98LMSVLRR172 pKa = 11.84TRR174 pKa = 11.84DD175 pKa = 3.4FSVLVVVNKK184 pKa = 10.17HH185 pKa = 5.09FADD188 pKa = 4.1PGFCALVHH196 pKa = 5.7EE197 pKa = 5.22RR198 pKa = 11.84YY199 pKa = 10.11DD200 pKa = 3.58VFVLDD205 pKa = 4.81EE206 pKa = 4.2SHH208 pKa = 6.98TYY210 pKa = 11.44NLMNLTLMTRR220 pKa = 11.84FLSFFPPRR228 pKa = 11.84ICYY231 pKa = 9.22FLSATPRR238 pKa = 11.84AANRR242 pKa = 11.84VYY244 pKa = 10.63CNRR247 pKa = 11.84VVNVTKK253 pKa = 10.38FSALRR258 pKa = 11.84KK259 pKa = 6.36TVRR262 pKa = 11.84VLSTFFPPFSNARR275 pKa = 11.84IRR277 pKa = 11.84EE278 pKa = 3.98FVQRR282 pKa = 11.84LDD284 pKa = 3.71SVHH287 pKa = 6.1NKK289 pKa = 7.37YY290 pKa = 10.49HH291 pKa = 6.98IYY293 pKa = 9.32TEE295 pKa = 4.02KK296 pKa = 11.01ALAEE300 pKa = 4.2DD301 pKa = 4.05APRR304 pKa = 11.84NASIVAVTAAAFEE317 pKa = 4.3HH318 pKa = 6.96RR319 pKa = 11.84DD320 pKa = 3.11AHH322 pKa = 6.24RR323 pKa = 11.84VLLITKK329 pKa = 9.37LRR331 pKa = 11.84SHH333 pKa = 6.59MLGMHH338 pKa = 7.0AALSARR344 pKa = 11.84LGAQHH349 pKa = 6.56VFLGDD354 pKa = 3.22AQSRR358 pKa = 11.84HH359 pKa = 4.85TPEE362 pKa = 3.8IVCEE366 pKa = 4.04LRR368 pKa = 11.84ASEE371 pKa = 4.28RR372 pKa = 11.84FVFVSTLFYY381 pKa = 10.98SGTGLDD387 pKa = 3.82IPHH390 pKa = 7.8LDD392 pKa = 3.53TLVVCHH398 pKa = 6.46AVMNHH403 pKa = 4.81MQAEE407 pKa = 4.16QLLGRR412 pKa = 11.84ICRR415 pKa = 11.84EE416 pKa = 3.73TEE418 pKa = 3.67QPRR421 pKa = 11.84RR422 pKa = 11.84TVYY425 pKa = 10.06VFPTTSVRR433 pKa = 11.84EE434 pKa = 4.12IKK436 pKa = 11.14NMVGLFAQRR445 pKa = 11.84FITLATQKK453 pKa = 10.93LGFTQEE459 pKa = 3.93RR460 pKa = 11.84EE461 pKa = 4.18QPEE464 pKa = 4.35DD465 pKa = 3.29AAVPASADD473 pKa = 3.71RR474 pKa = 11.84PASADD479 pKa = 2.88KK480 pKa = 10.48RR481 pKa = 11.84APADD485 pKa = 3.73RR486 pKa = 11.84PAPADD491 pKa = 3.89RR492 pKa = 11.84PASADD497 pKa = 2.88KK498 pKa = 10.48RR499 pKa = 11.84APADD503 pKa = 3.91RR504 pKa = 11.84PASADD509 pKa = 2.88KK510 pKa = 10.48RR511 pKa = 11.84APADD515 pKa = 3.91RR516 pKa = 11.84PASADD521 pKa = 2.88KK522 pKa = 10.48RR523 pKa = 11.84APADD527 pKa = 3.91RR528 pKa = 11.84PASADD533 pKa = 2.88KK534 pKa = 10.48RR535 pKa = 11.84APADD539 pKa = 3.83RR540 pKa = 11.84PASADD545 pKa = 3.84RR546 pKa = 11.84TAHH549 pKa = 6.39AYY551 pKa = 7.93WRR553 pKa = 11.84ANGPCCSNRR562 pKa = 11.84FASADD567 pKa = 3.44NKK569 pKa = 10.58KK570 pKa = 9.56PALSISRR577 pKa = 11.84ASGGDD582 pKa = 3.29NAVEE586 pKa = 4.53LGHH589 pKa = 6.79KK590 pKa = 9.23PGPGPGTHH598 pKa = 6.84ASLRR602 pKa = 11.84SVRR605 pKa = 11.84KK606 pKa = 9.23HH607 pKa = 5.29GRR609 pKa = 11.84SLACEE614 pKa = 4.32LAQGTQCRR622 pKa = 11.84SARR625 pKa = 11.84RR626 pKa = 11.84AVQRR630 pKa = 11.84VGPGRR635 pKa = 11.84GHH637 pKa = 6.41AQRR640 pKa = 11.84AGLQLAHH647 pKa = 7.24IGEE650 pKa = 4.37EE651 pKa = 4.47CEE653 pKa = 4.04ARR655 pKa = 11.84LAHH658 pKa = 6.65GARR661 pKa = 11.84GARR664 pKa = 11.84VRR666 pKa = 11.84GRR668 pKa = 11.84LLHH671 pKa = 6.29GGTRR675 pKa = 11.84LALAPPLLGSGHH687 pKa = 5.13VHH689 pKa = 7.34LDD691 pKa = 3.23AEE693 pKa = 4.67KK694 pKa = 10.81
MM1 pKa = 7.52SGFLAMDD8 pKa = 4.27DD9 pKa = 4.81KK10 pKa = 11.63LYY12 pKa = 10.84TDD14 pKa = 4.18LKK16 pKa = 10.3KK17 pKa = 10.89LVGAQPLYY25 pKa = 11.05LFTARR30 pKa = 11.84GDD32 pKa = 3.79FVEE35 pKa = 4.28VARR38 pKa = 11.84RR39 pKa = 11.84SEE41 pKa = 3.69FRR43 pKa = 11.84FRR45 pKa = 11.84VPAGFFAGEE54 pKa = 4.25GVALRR59 pKa = 11.84GQQFPAGSARR69 pKa = 11.84MPDD72 pKa = 3.49AAHH75 pKa = 6.56LRR77 pKa = 11.84LPRR80 pKa = 11.84LYY82 pKa = 9.75PAQQQVVAEE91 pKa = 4.2LRR93 pKa = 11.84AIAKK97 pKa = 9.8ACARR101 pKa = 11.84EE102 pKa = 3.88QRR104 pKa = 11.84PLYY107 pKa = 8.95VTLHH111 pKa = 6.28LACGFGKK118 pKa = 8.59TVTASYY124 pKa = 11.43LLGTHH129 pKa = 6.23RR130 pKa = 11.84RR131 pKa = 11.84RR132 pKa = 11.84AVVCVPTKK140 pKa = 10.3MLLAQWGAVLAALRR154 pKa = 11.84VSVYY158 pKa = 10.4VSGEE162 pKa = 4.04GVRR165 pKa = 11.84KK166 pKa = 9.98LMSVLRR172 pKa = 11.84TRR174 pKa = 11.84DD175 pKa = 3.4FSVLVVVNKK184 pKa = 10.17HH185 pKa = 5.09FADD188 pKa = 4.1PGFCALVHH196 pKa = 5.7EE197 pKa = 5.22RR198 pKa = 11.84YY199 pKa = 10.11DD200 pKa = 3.58VFVLDD205 pKa = 4.81EE206 pKa = 4.2SHH208 pKa = 6.98TYY210 pKa = 11.44NLMNLTLMTRR220 pKa = 11.84FLSFFPPRR228 pKa = 11.84ICYY231 pKa = 9.22FLSATPRR238 pKa = 11.84AANRR242 pKa = 11.84VYY244 pKa = 10.63CNRR247 pKa = 11.84VVNVTKK253 pKa = 10.38FSALRR258 pKa = 11.84KK259 pKa = 6.36TVRR262 pKa = 11.84VLSTFFPPFSNARR275 pKa = 11.84IRR277 pKa = 11.84EE278 pKa = 3.98FVQRR282 pKa = 11.84LDD284 pKa = 3.71SVHH287 pKa = 6.1NKK289 pKa = 7.37YY290 pKa = 10.49HH291 pKa = 6.98IYY293 pKa = 9.32TEE295 pKa = 4.02KK296 pKa = 11.01ALAEE300 pKa = 4.2DD301 pKa = 4.05APRR304 pKa = 11.84NASIVAVTAAAFEE317 pKa = 4.3HH318 pKa = 6.96RR319 pKa = 11.84DD320 pKa = 3.11AHH322 pKa = 6.24RR323 pKa = 11.84VLLITKK329 pKa = 9.37LRR331 pKa = 11.84SHH333 pKa = 6.59MLGMHH338 pKa = 7.0AALSARR344 pKa = 11.84LGAQHH349 pKa = 6.56VFLGDD354 pKa = 3.22AQSRR358 pKa = 11.84HH359 pKa = 4.85TPEE362 pKa = 3.8IVCEE366 pKa = 4.04LRR368 pKa = 11.84ASEE371 pKa = 4.28RR372 pKa = 11.84FVFVSTLFYY381 pKa = 10.98SGTGLDD387 pKa = 3.82IPHH390 pKa = 7.8LDD392 pKa = 3.53TLVVCHH398 pKa = 6.46AVMNHH403 pKa = 4.81MQAEE407 pKa = 4.16QLLGRR412 pKa = 11.84ICRR415 pKa = 11.84EE416 pKa = 3.73TEE418 pKa = 3.67QPRR421 pKa = 11.84RR422 pKa = 11.84TVYY425 pKa = 10.06VFPTTSVRR433 pKa = 11.84EE434 pKa = 4.12IKK436 pKa = 11.14NMVGLFAQRR445 pKa = 11.84FITLATQKK453 pKa = 10.93LGFTQEE459 pKa = 3.93RR460 pKa = 11.84EE461 pKa = 4.18QPEE464 pKa = 4.35DD465 pKa = 3.29AAVPASADD473 pKa = 3.71RR474 pKa = 11.84PASADD479 pKa = 2.88KK480 pKa = 10.48RR481 pKa = 11.84APADD485 pKa = 3.73RR486 pKa = 11.84PAPADD491 pKa = 3.89RR492 pKa = 11.84PASADD497 pKa = 2.88KK498 pKa = 10.48RR499 pKa = 11.84APADD503 pKa = 3.91RR504 pKa = 11.84PASADD509 pKa = 2.88KK510 pKa = 10.48RR511 pKa = 11.84APADD515 pKa = 3.91RR516 pKa = 11.84PASADD521 pKa = 2.88KK522 pKa = 10.48RR523 pKa = 11.84APADD527 pKa = 3.91RR528 pKa = 11.84PASADD533 pKa = 2.88KK534 pKa = 10.48RR535 pKa = 11.84APADD539 pKa = 3.83RR540 pKa = 11.84PASADD545 pKa = 3.84RR546 pKa = 11.84TAHH549 pKa = 6.39AYY551 pKa = 7.93WRR553 pKa = 11.84ANGPCCSNRR562 pKa = 11.84FASADD567 pKa = 3.44NKK569 pKa = 10.58KK570 pKa = 9.56PALSISRR577 pKa = 11.84ASGGDD582 pKa = 3.29NAVEE586 pKa = 4.53LGHH589 pKa = 6.79KK590 pKa = 9.23PGPGPGTHH598 pKa = 6.84ASLRR602 pKa = 11.84SVRR605 pKa = 11.84KK606 pKa = 9.23HH607 pKa = 5.29GRR609 pKa = 11.84SLACEE614 pKa = 4.32LAQGTQCRR622 pKa = 11.84SARR625 pKa = 11.84RR626 pKa = 11.84AVQRR630 pKa = 11.84VGPGRR635 pKa = 11.84GHH637 pKa = 6.41AQRR640 pKa = 11.84AGLQLAHH647 pKa = 7.24IGEE650 pKa = 4.37EE651 pKa = 4.47CEE653 pKa = 4.04ARR655 pKa = 11.84LAHH658 pKa = 6.65GARR661 pKa = 11.84GARR664 pKa = 11.84VRR666 pKa = 11.84GRR668 pKa = 11.84LLHH671 pKa = 6.29GGTRR675 pKa = 11.84LALAPPLLGSGHH687 pKa = 5.13VHH689 pKa = 7.34LDD691 pKa = 3.23AEE693 pKa = 4.67KK694 pKa = 10.81
Molecular weight: 76.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
54465 |
53 |
2133 |
334.1 |
37.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.818 ± 0.376 | 2.701 ± 0.14 |
4.785 ± 0.124 | 5.956 ± 0.143 |
4.17 ± 0.195 | 5.587 ± 0.195 |
2.745 ± 0.097 | 3.072 ± 0.23 |
2.998 ± 0.192 | 11.079 ± 0.206 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.187 ± 0.076 | 2.987 ± 0.162 |
5.473 ± 0.194 | 3.028 ± 0.165 |
8.815 ± 0.236 | 6.942 ± 0.186 |
4.966 ± 0.167 | 7.73 ± 0.154 |
0.736 ± 0.048 | 3.226 ± 0.112 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
