
Tentweb spider associated circular virus 1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.02
Get precalculated fractions of proteins
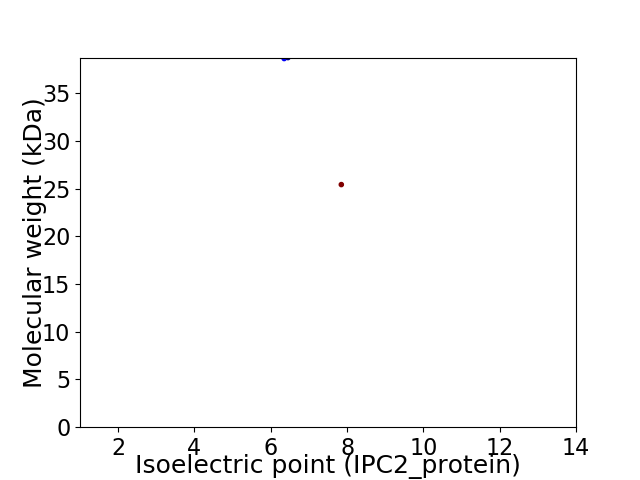
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BPD1|A0A346BPD1_9VIRU Putative capsid protein OS=Tentweb spider associated circular virus 1 OX=2293307 PE=4 SV=1
MM1 pKa = 7.56PRR3 pKa = 11.84QARR6 pKa = 11.84VKK8 pKa = 9.72CWCFTLNNYY17 pKa = 8.59TEE19 pKa = 5.57DD20 pKa = 3.6EE21 pKa = 4.41FLTLQRR27 pKa = 11.84TLEE30 pKa = 4.1LQKK33 pKa = 9.86PTFAIIAKK41 pKa = 9.72EE42 pKa = 3.99VGDD45 pKa = 3.92NQTPHH50 pKa = 5.83LQGYY54 pKa = 7.17VHH56 pKa = 6.33LTKK59 pKa = 10.07RR60 pKa = 11.84TVFSDD65 pKa = 3.19VKK67 pKa = 10.53KK68 pKa = 10.16ILGIRR73 pKa = 11.84AHH75 pKa = 6.68IEE77 pKa = 3.55QARR80 pKa = 11.84GSDD83 pKa = 3.58QDD85 pKa = 3.77SLLYY89 pKa = 10.36CSKK92 pKa = 10.61QDD94 pKa = 3.73PNPFIIGAPDD104 pKa = 3.78LKK106 pKa = 10.98SGGSAKK112 pKa = 10.32RR113 pKa = 11.84IALHH117 pKa = 6.31RR118 pKa = 11.84AASEE122 pKa = 4.15WIAGVTGGEE131 pKa = 4.51SIFHH135 pKa = 6.34QFTKK139 pKa = 10.82SPDD142 pKa = 2.97SGMAYY147 pKa = 9.93LRR149 pKa = 11.84HH150 pKa = 6.14FKK152 pKa = 10.55AIEE155 pKa = 4.25NIVKK159 pKa = 10.14DD160 pKa = 3.38KK161 pKa = 11.19RR162 pKa = 11.84RR163 pKa = 11.84IHH165 pKa = 7.41SMDD168 pKa = 4.14KK169 pKa = 10.38IRR171 pKa = 11.84EE172 pKa = 4.12EE173 pKa = 4.15YY174 pKa = 10.29TNATLRR180 pKa = 11.84IYY182 pKa = 8.81QARR185 pKa = 11.84IDD187 pKa = 4.1RR188 pKa = 11.84MVKK191 pKa = 10.44LPPDD195 pKa = 3.76PRR197 pKa = 11.84TIHH200 pKa = 5.76WFYY203 pKa = 11.4DD204 pKa = 3.31EE205 pKa = 4.99RR206 pKa = 11.84GNTGKK211 pKa = 10.06SWMTNWLMLNHH222 pKa = 6.82GAICFSNGKK231 pKa = 9.35SADD234 pKa = 2.92IAYY237 pKa = 9.37AYY239 pKa = 10.18SGEE242 pKa = 4.23RR243 pKa = 11.84IVVFDD248 pKa = 4.66FTRR251 pKa = 11.84SMQDD255 pKa = 2.91VLNYY259 pKa = 9.88QVIEE263 pKa = 3.95EE264 pKa = 4.57LKK266 pKa = 10.02NGRR269 pKa = 11.84IFSPKK274 pKa = 10.02YY275 pKa = 9.73EE276 pKa = 4.37SCCKK280 pKa = 10.21LFPRR284 pKa = 11.84PHH286 pKa = 6.76VICFANWLPDD296 pKa = 3.79YY297 pKa = 11.17GKK299 pKa = 9.95MSADD303 pKa = 2.79RR304 pKa = 11.84WDD306 pKa = 3.01IHH308 pKa = 6.91EE309 pKa = 5.55ILDD312 pKa = 3.87EE313 pKa = 4.65DD314 pKa = 4.98CEE316 pKa = 4.43QEE318 pKa = 3.83TDD320 pKa = 3.18QDD322 pKa = 3.91QVEE325 pKa = 4.36LEE327 pKa = 3.91NFIFEE332 pKa = 4.46KK333 pKa = 10.92
MM1 pKa = 7.56PRR3 pKa = 11.84QARR6 pKa = 11.84VKK8 pKa = 9.72CWCFTLNNYY17 pKa = 8.59TEE19 pKa = 5.57DD20 pKa = 3.6EE21 pKa = 4.41FLTLQRR27 pKa = 11.84TLEE30 pKa = 4.1LQKK33 pKa = 9.86PTFAIIAKK41 pKa = 9.72EE42 pKa = 3.99VGDD45 pKa = 3.92NQTPHH50 pKa = 5.83LQGYY54 pKa = 7.17VHH56 pKa = 6.33LTKK59 pKa = 10.07RR60 pKa = 11.84TVFSDD65 pKa = 3.19VKK67 pKa = 10.53KK68 pKa = 10.16ILGIRR73 pKa = 11.84AHH75 pKa = 6.68IEE77 pKa = 3.55QARR80 pKa = 11.84GSDD83 pKa = 3.58QDD85 pKa = 3.77SLLYY89 pKa = 10.36CSKK92 pKa = 10.61QDD94 pKa = 3.73PNPFIIGAPDD104 pKa = 3.78LKK106 pKa = 10.98SGGSAKK112 pKa = 10.32RR113 pKa = 11.84IALHH117 pKa = 6.31RR118 pKa = 11.84AASEE122 pKa = 4.15WIAGVTGGEE131 pKa = 4.51SIFHH135 pKa = 6.34QFTKK139 pKa = 10.82SPDD142 pKa = 2.97SGMAYY147 pKa = 9.93LRR149 pKa = 11.84HH150 pKa = 6.14FKK152 pKa = 10.55AIEE155 pKa = 4.25NIVKK159 pKa = 10.14DD160 pKa = 3.38KK161 pKa = 11.19RR162 pKa = 11.84RR163 pKa = 11.84IHH165 pKa = 7.41SMDD168 pKa = 4.14KK169 pKa = 10.38IRR171 pKa = 11.84EE172 pKa = 4.12EE173 pKa = 4.15YY174 pKa = 10.29TNATLRR180 pKa = 11.84IYY182 pKa = 8.81QARR185 pKa = 11.84IDD187 pKa = 4.1RR188 pKa = 11.84MVKK191 pKa = 10.44LPPDD195 pKa = 3.76PRR197 pKa = 11.84TIHH200 pKa = 5.76WFYY203 pKa = 11.4DD204 pKa = 3.31EE205 pKa = 4.99RR206 pKa = 11.84GNTGKK211 pKa = 10.06SWMTNWLMLNHH222 pKa = 6.82GAICFSNGKK231 pKa = 9.35SADD234 pKa = 2.92IAYY237 pKa = 9.37AYY239 pKa = 10.18SGEE242 pKa = 4.23RR243 pKa = 11.84IVVFDD248 pKa = 4.66FTRR251 pKa = 11.84SMQDD255 pKa = 2.91VLNYY259 pKa = 9.88QVIEE263 pKa = 3.95EE264 pKa = 4.57LKK266 pKa = 10.02NGRR269 pKa = 11.84IFSPKK274 pKa = 10.02YY275 pKa = 9.73EE276 pKa = 4.37SCCKK280 pKa = 10.21LFPRR284 pKa = 11.84PHH286 pKa = 6.76VICFANWLPDD296 pKa = 3.79YY297 pKa = 11.17GKK299 pKa = 9.95MSADD303 pKa = 2.79RR304 pKa = 11.84WDD306 pKa = 3.01IHH308 pKa = 6.91EE309 pKa = 5.55ILDD312 pKa = 3.87EE313 pKa = 4.65DD314 pKa = 4.98CEE316 pKa = 4.43QEE318 pKa = 3.83TDD320 pKa = 3.18QDD322 pKa = 3.91QVEE325 pKa = 4.36LEE327 pKa = 3.91NFIFEE332 pKa = 4.46KK333 pKa = 10.92
Molecular weight: 38.61 kDa
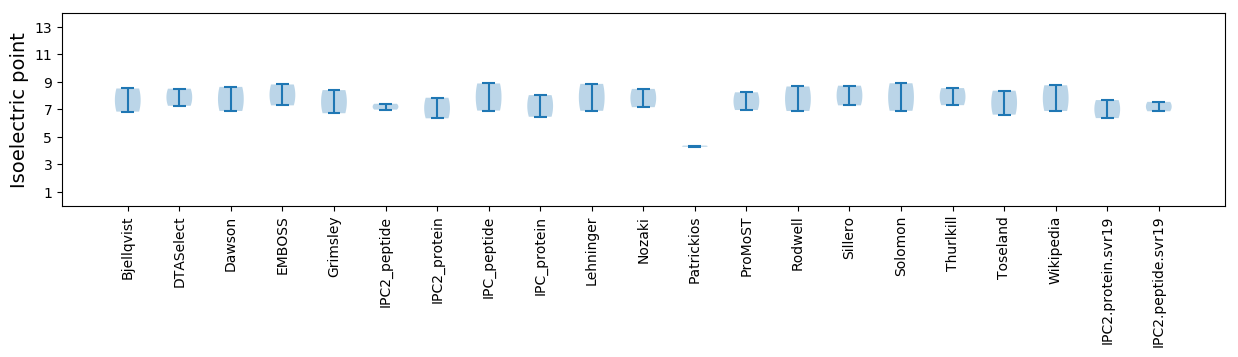
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BPD1|A0A346BPD1_9VIRU Putative capsid protein OS=Tentweb spider associated circular virus 1 OX=2293307 PE=4 SV=1
MM1 pKa = 8.03DD2 pKa = 4.48EE3 pKa = 3.87NVRR6 pKa = 11.84RR7 pKa = 11.84ILHH10 pKa = 6.88DD11 pKa = 4.61IIQQRR16 pKa = 11.84EE17 pKa = 3.68DD18 pKa = 3.57GYY20 pKa = 11.05RR21 pKa = 11.84SGMSAAIRR29 pKa = 11.84GIDD32 pKa = 3.66QSWSIPATGPFTTKK46 pKa = 9.79IRR48 pKa = 11.84FSLGDD53 pKa = 3.7LMKK56 pKa = 10.89SSATQYY62 pKa = 10.89SEE64 pKa = 5.84EE65 pKa = 4.14YY66 pKa = 8.24WQLEE70 pKa = 4.3GYY72 pKa = 7.88YY73 pKa = 10.23TIFDD77 pKa = 3.44QVKK80 pKa = 8.0VVKK83 pKa = 10.93VMFEE87 pKa = 3.79YY88 pKa = 10.4WLTDD92 pKa = 3.42DD93 pKa = 5.88DD94 pKa = 6.21VEE96 pKa = 4.58TTPNAAFVRR105 pKa = 11.84QVYY108 pKa = 10.47SYY110 pKa = 11.45DD111 pKa = 3.69PDD113 pKa = 3.9SADD116 pKa = 5.88GNMTWDD122 pKa = 3.41NVIRR126 pKa = 11.84VPDD129 pKa = 3.73HH130 pKa = 5.86AHH132 pKa = 7.19RR133 pKa = 11.84ILKK136 pKa = 10.02PGVIYY141 pKa = 8.66RR142 pKa = 11.84TSLKK146 pKa = 10.57PKK148 pKa = 10.04FAVQLATPNGTFKK161 pKa = 10.94GYY163 pKa = 10.44SKK165 pKa = 11.33NMWIDD170 pKa = 3.67CGDD173 pKa = 3.33VAGYY177 pKa = 9.73DD178 pKa = 3.43QAKK181 pKa = 10.1SVLGSVNAIHH191 pKa = 6.4MAFKK195 pKa = 10.78GPEE198 pKa = 3.7EE199 pKa = 4.34LGAIGYY205 pKa = 9.03RR206 pKa = 11.84RR207 pKa = 11.84TLFLKK212 pKa = 10.09FRR214 pKa = 11.84KK215 pKa = 9.15RR216 pKa = 11.84RR217 pKa = 11.84QGAVVNN223 pKa = 4.41
MM1 pKa = 8.03DD2 pKa = 4.48EE3 pKa = 3.87NVRR6 pKa = 11.84RR7 pKa = 11.84ILHH10 pKa = 6.88DD11 pKa = 4.61IIQQRR16 pKa = 11.84EE17 pKa = 3.68DD18 pKa = 3.57GYY20 pKa = 11.05RR21 pKa = 11.84SGMSAAIRR29 pKa = 11.84GIDD32 pKa = 3.66QSWSIPATGPFTTKK46 pKa = 9.79IRR48 pKa = 11.84FSLGDD53 pKa = 3.7LMKK56 pKa = 10.89SSATQYY62 pKa = 10.89SEE64 pKa = 5.84EE65 pKa = 4.14YY66 pKa = 8.24WQLEE70 pKa = 4.3GYY72 pKa = 7.88YY73 pKa = 10.23TIFDD77 pKa = 3.44QVKK80 pKa = 8.0VVKK83 pKa = 10.93VMFEE87 pKa = 3.79YY88 pKa = 10.4WLTDD92 pKa = 3.42DD93 pKa = 5.88DD94 pKa = 6.21VEE96 pKa = 4.58TTPNAAFVRR105 pKa = 11.84QVYY108 pKa = 10.47SYY110 pKa = 11.45DD111 pKa = 3.69PDD113 pKa = 3.9SADD116 pKa = 5.88GNMTWDD122 pKa = 3.41NVIRR126 pKa = 11.84VPDD129 pKa = 3.73HH130 pKa = 5.86AHH132 pKa = 7.19RR133 pKa = 11.84ILKK136 pKa = 10.02PGVIYY141 pKa = 8.66RR142 pKa = 11.84TSLKK146 pKa = 10.57PKK148 pKa = 10.04FAVQLATPNGTFKK161 pKa = 10.94GYY163 pKa = 10.44SKK165 pKa = 11.33NMWIDD170 pKa = 3.67CGDD173 pKa = 3.33VAGYY177 pKa = 9.73DD178 pKa = 3.43QAKK181 pKa = 10.1SVLGSVNAIHH191 pKa = 6.4MAFKK195 pKa = 10.78GPEE198 pKa = 3.7EE199 pKa = 4.34LGAIGYY205 pKa = 9.03RR206 pKa = 11.84RR207 pKa = 11.84TLFLKK212 pKa = 10.09FRR214 pKa = 11.84KK215 pKa = 9.15RR216 pKa = 11.84RR217 pKa = 11.84QGAVVNN223 pKa = 4.41
Molecular weight: 25.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
556 |
223 |
333 |
278.0 |
32.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.655 ± 0.29 | 1.619 ± 0.653 |
7.194 ± 0.239 | 5.576 ± 0.859 |
4.856 ± 0.207 | 6.295 ± 0.741 |
2.698 ± 0.504 | 7.194 ± 0.511 |
6.295 ± 0.26 | 6.295 ± 0.51 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.698 ± 0.246 | 3.957 ± 0.206 |
4.137 ± 0.056 | 4.496 ± 0.007 |
6.655 ± 0.04 | 5.935 ± 0.191 |
5.396 ± 0.242 | 5.576 ± 1.142 |
2.158 ± 0.047 | 4.317 ± 0.594 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
