
Actinobacteria bacterium OV320
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; unclassified Actinomycetia
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
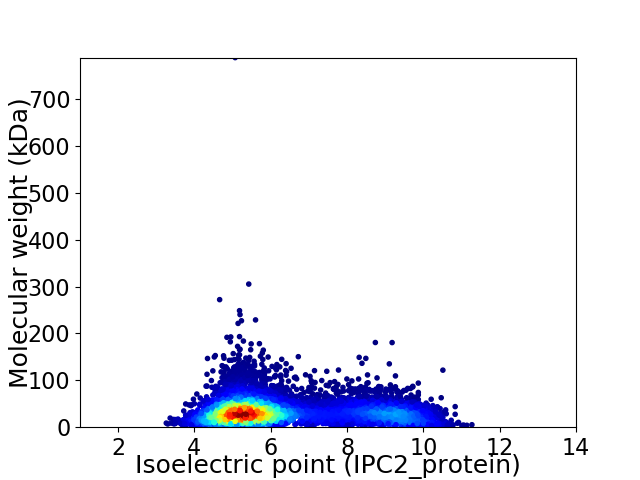
Virtual 2D-PAGE plot for 9056 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
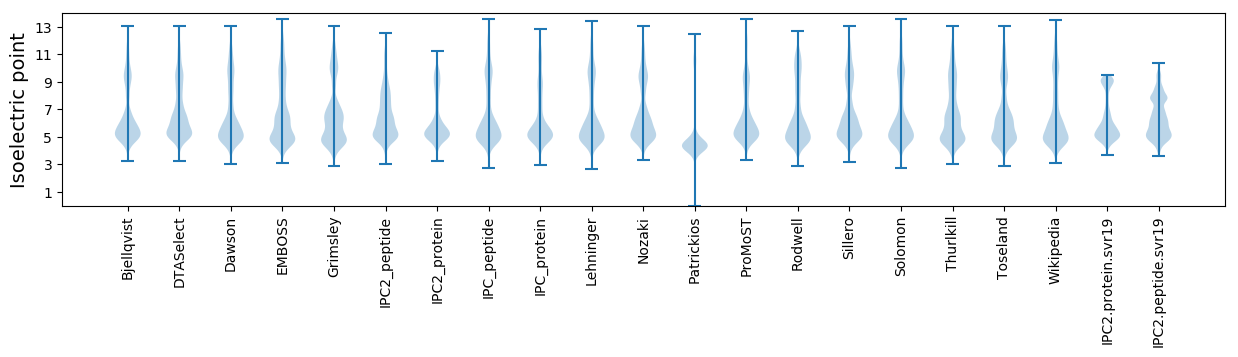
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0N1GK75|A0A0N1GK75_9ACTN ABC-type transporter integral membrane subunit OS=Actinobacteria bacterium OV320 OX=1592329 GN=OV320_7245 PE=3 SV=1
MM1 pKa = 7.81SIPPPPGPHH10 pKa = 5.88QPQDD14 pKa = 3.84PYY16 pKa = 11.35QPPQPQSPYY25 pKa = 9.04PQDD28 pKa = 3.62PFAPPPQDD36 pKa = 3.86PYY38 pKa = 11.7AQGPYY43 pKa = 10.17GPQGPQAPMGPGPMGPGPMGPGPGPMGPYY72 pKa = 8.93PHH74 pKa = 7.58PSYY77 pKa = 10.58PQAPYY82 pKa = 10.61GAVPYY87 pKa = 8.8PVWGQGYY94 pKa = 9.06SPYY97 pKa = 10.63GRR99 pKa = 11.84PSPVNGVAIAALVLGLLCFLPGVGLLLGIIALVQIRR135 pKa = 11.84KK136 pKa = 9.57RR137 pKa = 11.84GEE139 pKa = 3.57RR140 pKa = 11.84GKK142 pKa = 11.14AMAIVGTVLSSVGLAVWVLALSTGGASDD170 pKa = 3.57FWDD173 pKa = 4.08GFKK176 pKa = 10.72EE177 pKa = 4.31GASGNSSFALAKK189 pKa = 10.13GDD191 pKa = 3.93CFDD194 pKa = 4.22VPGEE198 pKa = 4.19TFDD201 pKa = 3.85EE202 pKa = 4.34DD203 pKa = 4.44VYY205 pKa = 11.81DD206 pKa = 4.06VDD208 pKa = 5.54EE209 pKa = 4.46VDD211 pKa = 4.93CSGEE215 pKa = 3.54HH216 pKa = 6.53DD217 pKa = 3.83AEE219 pKa = 4.55VFASVPLTGDD229 pKa = 3.45SFPGDD234 pKa = 3.42DD235 pKa = 4.44HH236 pKa = 6.23VTDD239 pKa = 3.76VADD242 pKa = 4.43DD243 pKa = 3.47KK244 pKa = 11.7CYY246 pKa = 10.12TLQDD250 pKa = 4.01AYY252 pKa = 11.05AMDD255 pKa = 4.25PWALTDD261 pKa = 4.07EE262 pKa = 4.14VDD264 pKa = 4.09IYY266 pKa = 11.52YY267 pKa = 7.57LTPTADD273 pKa = 2.79SWSWGDD279 pKa = 4.3RR280 pKa = 11.84EE281 pKa = 4.37ITCVFANVDD290 pKa = 3.61EE291 pKa = 5.11KK292 pKa = 11.33GTLTGSLRR300 pKa = 11.84ADD302 pKa = 3.72ASTLDD307 pKa = 3.66PDD309 pKa = 3.55QFAFLKK315 pKa = 10.93AMAAVDD321 pKa = 3.68DD322 pKa = 4.28VLFEE326 pKa = 4.23EE327 pKa = 4.77PEE329 pKa = 4.1EE330 pKa = 4.06YY331 pKa = 10.71AEE333 pKa = 5.31EE334 pKa = 4.41DD335 pKa = 3.52LASNRR340 pKa = 11.84TWAGDD345 pKa = 3.39TGEE348 pKa = 4.34VTGNQADD355 pKa = 3.62QLGSHH360 pKa = 6.4LWEE363 pKa = 4.59EE364 pKa = 4.63PDD366 pKa = 4.41RR367 pKa = 11.84EE368 pKa = 4.36APLAALVKK376 pKa = 10.64DD377 pKa = 3.43MRR379 pKa = 11.84TASKK383 pKa = 10.06EE384 pKa = 3.39WAAAAKK390 pKa = 10.22AADD393 pKa = 3.39ADD395 pKa = 4.13GFYY398 pKa = 10.85EE399 pKa = 5.18HH400 pKa = 7.27YY401 pKa = 10.55DD402 pKa = 3.22KK403 pKa = 11.04AYY405 pKa = 9.93KK406 pKa = 10.33YY407 pKa = 10.85VDD409 pKa = 3.18GATTVTARR417 pKa = 11.84KK418 pKa = 9.59ALGLATTVPTYY429 pKa = 11.03DD430 pKa = 3.86EE431 pKa = 5.75DD432 pKa = 4.39SDD434 pKa = 4.15SGSGGTGGGDD444 pKa = 3.25SDD446 pKa = 5.04SGLDD450 pKa = 3.39VV451 pKa = 3.72
MM1 pKa = 7.81SIPPPPGPHH10 pKa = 5.88QPQDD14 pKa = 3.84PYY16 pKa = 11.35QPPQPQSPYY25 pKa = 9.04PQDD28 pKa = 3.62PFAPPPQDD36 pKa = 3.86PYY38 pKa = 11.7AQGPYY43 pKa = 10.17GPQGPQAPMGPGPMGPGPMGPGPGPMGPYY72 pKa = 8.93PHH74 pKa = 7.58PSYY77 pKa = 10.58PQAPYY82 pKa = 10.61GAVPYY87 pKa = 8.8PVWGQGYY94 pKa = 9.06SPYY97 pKa = 10.63GRR99 pKa = 11.84PSPVNGVAIAALVLGLLCFLPGVGLLLGIIALVQIRR135 pKa = 11.84KK136 pKa = 9.57RR137 pKa = 11.84GEE139 pKa = 3.57RR140 pKa = 11.84GKK142 pKa = 11.14AMAIVGTVLSSVGLAVWVLALSTGGASDD170 pKa = 3.57FWDD173 pKa = 4.08GFKK176 pKa = 10.72EE177 pKa = 4.31GASGNSSFALAKK189 pKa = 10.13GDD191 pKa = 3.93CFDD194 pKa = 4.22VPGEE198 pKa = 4.19TFDD201 pKa = 3.85EE202 pKa = 4.34DD203 pKa = 4.44VYY205 pKa = 11.81DD206 pKa = 4.06VDD208 pKa = 5.54EE209 pKa = 4.46VDD211 pKa = 4.93CSGEE215 pKa = 3.54HH216 pKa = 6.53DD217 pKa = 3.83AEE219 pKa = 4.55VFASVPLTGDD229 pKa = 3.45SFPGDD234 pKa = 3.42DD235 pKa = 4.44HH236 pKa = 6.23VTDD239 pKa = 3.76VADD242 pKa = 4.43DD243 pKa = 3.47KK244 pKa = 11.7CYY246 pKa = 10.12TLQDD250 pKa = 4.01AYY252 pKa = 11.05AMDD255 pKa = 4.25PWALTDD261 pKa = 4.07EE262 pKa = 4.14VDD264 pKa = 4.09IYY266 pKa = 11.52YY267 pKa = 7.57LTPTADD273 pKa = 2.79SWSWGDD279 pKa = 4.3RR280 pKa = 11.84EE281 pKa = 4.37ITCVFANVDD290 pKa = 3.61EE291 pKa = 5.11KK292 pKa = 11.33GTLTGSLRR300 pKa = 11.84ADD302 pKa = 3.72ASTLDD307 pKa = 3.66PDD309 pKa = 3.55QFAFLKK315 pKa = 10.93AMAAVDD321 pKa = 3.68DD322 pKa = 4.28VLFEE326 pKa = 4.23EE327 pKa = 4.77PEE329 pKa = 4.1EE330 pKa = 4.06YY331 pKa = 10.71AEE333 pKa = 5.31EE334 pKa = 4.41DD335 pKa = 3.52LASNRR340 pKa = 11.84TWAGDD345 pKa = 3.39TGEE348 pKa = 4.34VTGNQADD355 pKa = 3.62QLGSHH360 pKa = 6.4LWEE363 pKa = 4.59EE364 pKa = 4.63PDD366 pKa = 4.41RR367 pKa = 11.84EE368 pKa = 4.36APLAALVKK376 pKa = 10.64DD377 pKa = 3.43MRR379 pKa = 11.84TASKK383 pKa = 10.06EE384 pKa = 3.39WAAAAKK390 pKa = 10.22AADD393 pKa = 3.39ADD395 pKa = 4.13GFYY398 pKa = 10.85EE399 pKa = 5.18HH400 pKa = 7.27YY401 pKa = 10.55DD402 pKa = 3.22KK403 pKa = 11.04AYY405 pKa = 9.93KK406 pKa = 10.33YY407 pKa = 10.85VDD409 pKa = 3.18GATTVTARR417 pKa = 11.84KK418 pKa = 9.59ALGLATTVPTYY429 pKa = 11.03DD430 pKa = 3.86EE431 pKa = 5.75DD432 pKa = 4.39SDD434 pKa = 4.15SGSGGTGGGDD444 pKa = 3.25SDD446 pKa = 5.04SGLDD450 pKa = 3.39VV451 pKa = 3.72
Molecular weight: 47.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0N1GRG6|A0A0N1GRG6_9ACTN Putative anti-sigma regulatory factor serine/threonine protein kinase OS=Actinobacteria bacterium OV320 OX=1592329 GN=OV320_6689 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILANRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 11.13GRR40 pKa = 11.84TNLSAA45 pKa = 4.57
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILANRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 11.13GRR40 pKa = 11.84TNLSAA45 pKa = 4.57
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2938333 |
30 |
7352 |
324.5 |
34.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.522 ± 0.045 | 0.781 ± 0.007 |
6.019 ± 0.021 | 5.622 ± 0.024 |
2.711 ± 0.015 | 9.455 ± 0.025 |
2.307 ± 0.012 | 3.11 ± 0.017 |
2.168 ± 0.021 | 10.237 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.681 ± 0.011 | 1.817 ± 0.017 |
6.104 ± 0.026 | 2.791 ± 0.016 |
7.973 ± 0.029 | 5.196 ± 0.027 |
6.358 ± 0.026 | 8.471 ± 0.025 |
1.544 ± 0.01 | 2.135 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
