
Desulfoluna spongiiphila
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobacteraceae; Desulfoluna
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
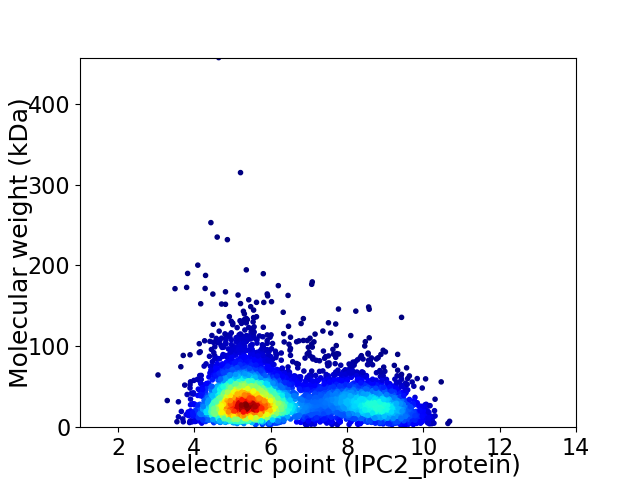
Virtual 2D-PAGE plot for 5238 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G5D6N5|A0A1G5D6N5_9DELT Long-chain fatty acid transport protein OS=Desulfoluna spongiiphila OX=419481 GN=SAMN05216233_10410 PE=3 SV=1
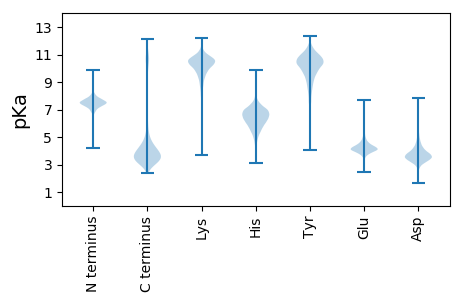
MM1 pKa = 7.98RR2 pKa = 11.84KK3 pKa = 8.87PLGPIGTLLCRR14 pKa = 11.84CLSYY18 pKa = 10.93LAGSPARR25 pKa = 11.84TGIRR29 pKa = 11.84GLSLLVALLALPCVASGMEE48 pKa = 4.31ALSEE52 pKa = 4.02HH53 pKa = 5.38QMKK56 pKa = 8.8GTTAQAGISIAMDD69 pKa = 3.45DD70 pKa = 3.6VTFYY74 pKa = 10.96HH75 pKa = 6.67HH76 pKa = 7.14VDD78 pKa = 3.25AFRR81 pKa = 11.84LTDD84 pKa = 4.08PDD86 pKa = 4.24DD87 pKa = 4.2PSGYY91 pKa = 9.22MEE93 pKa = 4.33FQDD96 pKa = 4.41IEE98 pKa = 4.53SLTTIDD104 pKa = 4.75VGTSDD109 pKa = 4.15VNGDD113 pKa = 3.48NTIGCITLDD122 pKa = 3.82LFTVSDD128 pKa = 3.76ATSPINGKK136 pKa = 8.77PLLYY140 pKa = 10.64LVGADD145 pKa = 2.86IDD147 pKa = 4.31YY148 pKa = 11.49NKK150 pKa = 10.63DD151 pKa = 3.22LTIGAINVCGTDD163 pKa = 2.89IGSAAVDD170 pKa = 3.76NYY172 pKa = 11.52AMPSFHH178 pKa = 7.25LYY180 pKa = 10.51LGAHH184 pKa = 6.82DD185 pKa = 4.99CGVDD189 pKa = 3.46LEE191 pKa = 4.62FGARR195 pKa = 11.84ITMDD199 pKa = 2.99QFSYY203 pKa = 9.23GTSPDD208 pKa = 3.52DD209 pKa = 3.43TLTLSGITLAGSFTDD224 pKa = 4.61NPADD228 pKa = 3.96TPSDD232 pKa = 3.54PSTWQAGGEE241 pKa = 4.15FVLGDD246 pKa = 3.57IASGNPLTIDD256 pKa = 3.3VATDD260 pKa = 3.7TTASWADD267 pKa = 3.68DD268 pKa = 3.74ADD270 pKa = 4.27GNPIPNPRR278 pKa = 11.84LGSGFIAINMPMEE291 pKa = 4.48GSLRR295 pKa = 11.84IEE297 pKa = 3.86NMGFGTNDD305 pKa = 3.31FGALALDD312 pKa = 4.74NIQAHH317 pKa = 6.57KK318 pKa = 10.54LYY320 pKa = 10.44IEE322 pKa = 4.11IPGRR326 pKa = 11.84GLGVPP331 pKa = 4.21
MM1 pKa = 7.98RR2 pKa = 11.84KK3 pKa = 8.87PLGPIGTLLCRR14 pKa = 11.84CLSYY18 pKa = 10.93LAGSPARR25 pKa = 11.84TGIRR29 pKa = 11.84GLSLLVALLALPCVASGMEE48 pKa = 4.31ALSEE52 pKa = 4.02HH53 pKa = 5.38QMKK56 pKa = 8.8GTTAQAGISIAMDD69 pKa = 3.45DD70 pKa = 3.6VTFYY74 pKa = 10.96HH75 pKa = 6.67HH76 pKa = 7.14VDD78 pKa = 3.25AFRR81 pKa = 11.84LTDD84 pKa = 4.08PDD86 pKa = 4.24DD87 pKa = 4.2PSGYY91 pKa = 9.22MEE93 pKa = 4.33FQDD96 pKa = 4.41IEE98 pKa = 4.53SLTTIDD104 pKa = 4.75VGTSDD109 pKa = 4.15VNGDD113 pKa = 3.48NTIGCITLDD122 pKa = 3.82LFTVSDD128 pKa = 3.76ATSPINGKK136 pKa = 8.77PLLYY140 pKa = 10.64LVGADD145 pKa = 2.86IDD147 pKa = 4.31YY148 pKa = 11.49NKK150 pKa = 10.63DD151 pKa = 3.22LTIGAINVCGTDD163 pKa = 2.89IGSAAVDD170 pKa = 3.76NYY172 pKa = 11.52AMPSFHH178 pKa = 7.25LYY180 pKa = 10.51LGAHH184 pKa = 6.82DD185 pKa = 4.99CGVDD189 pKa = 3.46LEE191 pKa = 4.62FGARR195 pKa = 11.84ITMDD199 pKa = 2.99QFSYY203 pKa = 9.23GTSPDD208 pKa = 3.52DD209 pKa = 3.43TLTLSGITLAGSFTDD224 pKa = 4.61NPADD228 pKa = 3.96TPSDD232 pKa = 3.54PSTWQAGGEE241 pKa = 4.15FVLGDD246 pKa = 3.57IASGNPLTIDD256 pKa = 3.3VATDD260 pKa = 3.7TTASWADD267 pKa = 3.68DD268 pKa = 3.74ADD270 pKa = 4.27GNPIPNPRR278 pKa = 11.84LGSGFIAINMPMEE291 pKa = 4.48GSLRR295 pKa = 11.84IEE297 pKa = 3.86NMGFGTNDD305 pKa = 3.31FGALALDD312 pKa = 4.74NIQAHH317 pKa = 6.57KK318 pKa = 10.54LYY320 pKa = 10.44IEE322 pKa = 4.11IPGRR326 pKa = 11.84GLGVPP331 pKa = 4.21
Molecular weight: 34.72 kDa
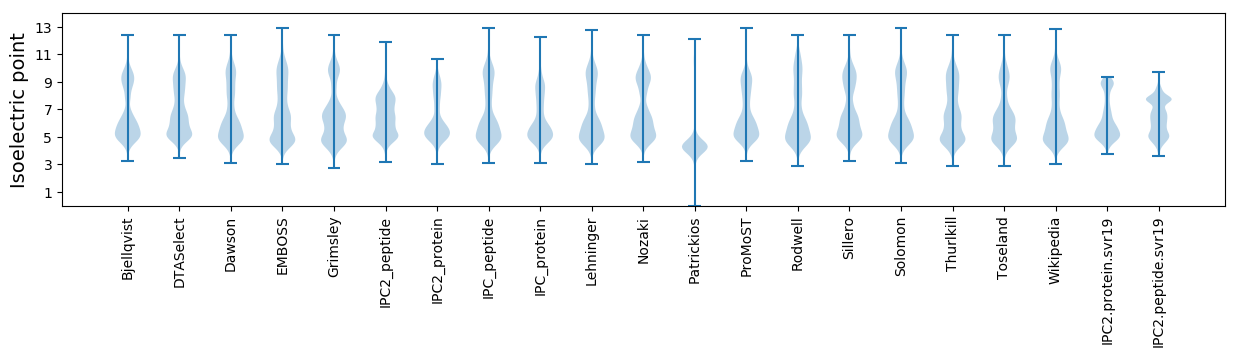
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G5ALR7|A0A1G5ALR7_9DELT Phosphoglycolate phosphatase OS=Desulfoluna spongiiphila OX=419481 GN=SAMN05216233_101285 PE=4 SV=1
MM1 pKa = 7.5RR2 pKa = 11.84WRR4 pKa = 11.84WLSALALMVRR14 pKa = 11.84RR15 pKa = 11.84LVYY18 pKa = 9.84PPRR21 pKa = 11.84CRR23 pKa = 11.84VCGVLLPWEE32 pKa = 4.17RR33 pKa = 11.84VGGSSSLHH41 pKa = 6.29SGDD44 pKa = 3.92LASMFHH50 pKa = 7.16GEE52 pKa = 4.36LCHH55 pKa = 7.23VMCATCRR62 pKa = 11.84AGVQEE67 pKa = 4.73PSGQRR72 pKa = 11.84CTACGQPLSGVVFDD86 pKa = 5.45PPLCGPCMDD95 pKa = 5.33PDD97 pKa = 3.89RR98 pKa = 11.84PLLRR102 pKa = 11.84MTALYY107 pKa = 10.52LHH109 pKa = 7.34DD110 pKa = 5.59GGPAMAVRR118 pKa = 11.84ALKK121 pKa = 10.49YY122 pKa = 9.77KK123 pKa = 10.41NKK125 pKa = 9.92RR126 pKa = 11.84ALAPPMGRR134 pKa = 11.84LLFARR139 pKa = 11.84ALRR142 pKa = 11.84AHH144 pKa = 7.28LDD146 pKa = 3.76GEE148 pKa = 4.35AKK150 pKa = 10.27RR151 pKa = 11.84WDD153 pKa = 3.38IDD155 pKa = 3.76LVVPVPLHH163 pKa = 5.63GKK165 pKa = 8.57RR166 pKa = 11.84LKK168 pKa = 10.92ARR170 pKa = 11.84GFNQASVLLAAFEE183 pKa = 4.46KK184 pKa = 10.57EE185 pKa = 3.51AGLRR189 pKa = 11.84VCHH192 pKa = 6.32RR193 pKa = 11.84LLRR196 pKa = 11.84RR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84PTTPQAGLSRR209 pKa = 11.84RR210 pKa = 11.84ARR212 pKa = 11.84LTNVKK217 pKa = 10.04GAFEE221 pKa = 4.26LAPGRR226 pKa = 11.84MVEE229 pKa = 4.08GLKK232 pKa = 10.65VLLVDD237 pKa = 4.96DD238 pKa = 5.09VYY240 pKa = 9.5TTGSTLAACAALLKK254 pKa = 10.5RR255 pKa = 11.84HH256 pKa = 5.92GATEE260 pKa = 3.98VSALVFARR268 pKa = 11.84RR269 pKa = 11.84DD270 pKa = 3.42MTHH273 pKa = 6.05
MM1 pKa = 7.5RR2 pKa = 11.84WRR4 pKa = 11.84WLSALALMVRR14 pKa = 11.84RR15 pKa = 11.84LVYY18 pKa = 9.84PPRR21 pKa = 11.84CRR23 pKa = 11.84VCGVLLPWEE32 pKa = 4.17RR33 pKa = 11.84VGGSSSLHH41 pKa = 6.29SGDD44 pKa = 3.92LASMFHH50 pKa = 7.16GEE52 pKa = 4.36LCHH55 pKa = 7.23VMCATCRR62 pKa = 11.84AGVQEE67 pKa = 4.73PSGQRR72 pKa = 11.84CTACGQPLSGVVFDD86 pKa = 5.45PPLCGPCMDD95 pKa = 5.33PDD97 pKa = 3.89RR98 pKa = 11.84PLLRR102 pKa = 11.84MTALYY107 pKa = 10.52LHH109 pKa = 7.34DD110 pKa = 5.59GGPAMAVRR118 pKa = 11.84ALKK121 pKa = 10.49YY122 pKa = 9.77KK123 pKa = 10.41NKK125 pKa = 9.92RR126 pKa = 11.84ALAPPMGRR134 pKa = 11.84LLFARR139 pKa = 11.84ALRR142 pKa = 11.84AHH144 pKa = 7.28LDD146 pKa = 3.76GEE148 pKa = 4.35AKK150 pKa = 10.27RR151 pKa = 11.84WDD153 pKa = 3.38IDD155 pKa = 3.76LVVPVPLHH163 pKa = 5.63GKK165 pKa = 8.57RR166 pKa = 11.84LKK168 pKa = 10.92ARR170 pKa = 11.84GFNQASVLLAAFEE183 pKa = 4.46KK184 pKa = 10.57EE185 pKa = 3.51AGLRR189 pKa = 11.84VCHH192 pKa = 6.32RR193 pKa = 11.84LLRR196 pKa = 11.84RR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84PTTPQAGLSRR209 pKa = 11.84RR210 pKa = 11.84ARR212 pKa = 11.84LTNVKK217 pKa = 10.04GAFEE221 pKa = 4.26LAPGRR226 pKa = 11.84MVEE229 pKa = 4.08GLKK232 pKa = 10.65VLLVDD237 pKa = 4.96DD238 pKa = 5.09VYY240 pKa = 9.5TTGSTLAACAALLKK254 pKa = 10.5RR255 pKa = 11.84HH256 pKa = 5.92GATEE260 pKa = 3.98VSALVFARR268 pKa = 11.84RR269 pKa = 11.84DD270 pKa = 3.42MTHH273 pKa = 6.05
Molecular weight: 30.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1765537 |
25 |
4215 |
337.1 |
37.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.365 ± 0.04 | 1.309 ± 0.015 |
5.541 ± 0.031 | 6.559 ± 0.033 |
4.171 ± 0.023 | 8.205 ± 0.03 |
2.204 ± 0.016 | 5.694 ± 0.032 |
4.939 ± 0.034 | 9.879 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.898 ± 0.018 | 3.122 ± 0.021 |
4.582 ± 0.023 | 2.684 ± 0.016 |
5.762 ± 0.029 | 5.924 ± 0.026 |
5.876 ± 0.028 | 7.391 ± 0.032 |
1.133 ± 0.014 | 2.762 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
