
Palaemonetes sp. common grass shrimp associated circular virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.18
Get precalculated fractions of proteins
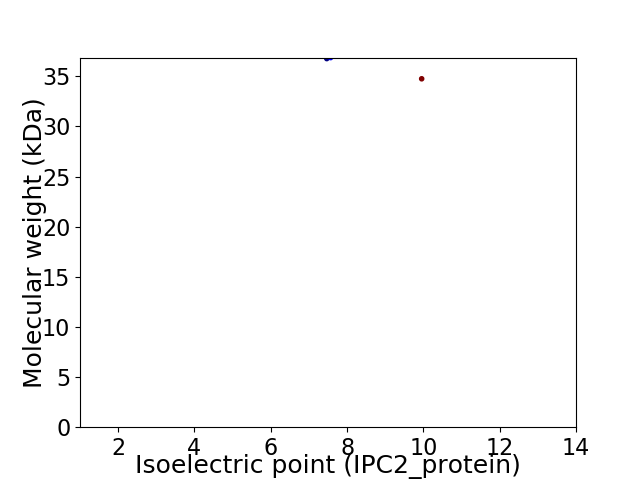
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1RL70|A0A0K1RL70_9CIRC Putative capsid protein OS=Palaemonetes sp. common grass shrimp associated circular virus OX=1692259 PE=4 SV=1
MM1 pKa = 7.33AAKK4 pKa = 9.89RR5 pKa = 11.84DD6 pKa = 3.86SAVKK10 pKa = 10.14RR11 pKa = 11.84WCFTLNNYY19 pKa = 8.28TDD21 pKa = 4.45ADD23 pKa = 4.03CEE25 pKa = 4.08ALKK28 pKa = 10.87EE29 pKa = 4.05KK30 pKa = 9.96LTTDD34 pKa = 3.18TCSRR38 pKa = 11.84AIVGKK43 pKa = 10.32EE44 pKa = 3.38KK45 pKa = 11.23GEE47 pKa = 4.05NGTPHH52 pKa = 6.39LQGFVSLKK60 pKa = 7.08TRR62 pKa = 11.84KK63 pKa = 9.48RR64 pKa = 11.84LSAMKK69 pKa = 9.67TFLSPRR75 pKa = 11.84YY76 pKa = 9.46HH77 pKa = 6.8FEE79 pKa = 3.75QAKK82 pKa = 8.4GTDD85 pKa = 3.66EE86 pKa = 4.65QNTEE90 pKa = 4.11YY91 pKa = 10.5CSKK94 pKa = 10.73EE95 pKa = 3.63GDD97 pKa = 3.64VLIDD101 pKa = 2.96VGEE104 pKa = 4.16NVKK107 pKa = 10.47GRR109 pKa = 11.84GDD111 pKa = 3.11KK112 pKa = 11.26GGGFNEE118 pKa = 4.3GANIKK123 pKa = 10.2RR124 pKa = 11.84VVYY127 pKa = 10.31HH128 pKa = 6.54LLEE131 pKa = 4.98GKK133 pKa = 10.08DD134 pKa = 4.06KK135 pKa = 11.01EE136 pKa = 5.41ALLQDD141 pKa = 3.74EE142 pKa = 4.97KK143 pKa = 11.32MLGAYY148 pKa = 7.45MKK150 pKa = 9.3YY151 pKa = 9.92KK152 pKa = 10.52RR153 pKa = 11.84SITEE157 pKa = 3.62MVSEE161 pKa = 4.38VKK163 pKa = 10.4SEE165 pKa = 4.06QEE167 pKa = 4.06TKK169 pKa = 10.44RR170 pKa = 11.84LKK172 pKa = 10.31QDD174 pKa = 3.37LDD176 pKa = 3.69NATLRR181 pKa = 11.84PWQRR185 pKa = 11.84EE186 pKa = 3.8LKK188 pKa = 10.05EE189 pKa = 4.05YY190 pKa = 9.91LTGDD194 pKa = 3.09PHH196 pKa = 6.94DD197 pKa = 4.34RR198 pKa = 11.84HH199 pKa = 7.2IIWYY203 pKa = 9.54IDD205 pKa = 3.35GQGNNGKK212 pKa = 9.11SWFAKK217 pKa = 9.65YY218 pKa = 9.85CVAEE222 pKa = 4.37LGAIRR227 pKa = 11.84LEE229 pKa = 4.2NAKK232 pKa = 8.62TADD235 pKa = 3.42LTHH238 pKa = 7.17AYY240 pKa = 9.89NGEE243 pKa = 3.94RR244 pKa = 11.84VVIFDD249 pKa = 4.88LSRR252 pKa = 11.84TTEE255 pKa = 3.75GHH257 pKa = 5.82FNYY260 pKa = 10.36GALEE264 pKa = 4.28SIKK267 pKa = 10.36NGCVFSSKK275 pKa = 10.46YY276 pKa = 10.42DD277 pKa = 3.4SRR279 pKa = 11.84QKK281 pKa = 9.99MFPIPHH287 pKa = 6.29VIVFANWGPDD297 pKa = 3.06RR298 pKa = 11.84SKK300 pKa = 10.92LSEE303 pKa = 4.33DD304 pKa = 2.84RR305 pKa = 11.84WVTKK309 pKa = 8.48TWSFEE314 pKa = 3.86QEE316 pKa = 4.04TFQTFF321 pKa = 2.92
MM1 pKa = 7.33AAKK4 pKa = 9.89RR5 pKa = 11.84DD6 pKa = 3.86SAVKK10 pKa = 10.14RR11 pKa = 11.84WCFTLNNYY19 pKa = 8.28TDD21 pKa = 4.45ADD23 pKa = 4.03CEE25 pKa = 4.08ALKK28 pKa = 10.87EE29 pKa = 4.05KK30 pKa = 9.96LTTDD34 pKa = 3.18TCSRR38 pKa = 11.84AIVGKK43 pKa = 10.32EE44 pKa = 3.38KK45 pKa = 11.23GEE47 pKa = 4.05NGTPHH52 pKa = 6.39LQGFVSLKK60 pKa = 7.08TRR62 pKa = 11.84KK63 pKa = 9.48RR64 pKa = 11.84LSAMKK69 pKa = 9.67TFLSPRR75 pKa = 11.84YY76 pKa = 9.46HH77 pKa = 6.8FEE79 pKa = 3.75QAKK82 pKa = 8.4GTDD85 pKa = 3.66EE86 pKa = 4.65QNTEE90 pKa = 4.11YY91 pKa = 10.5CSKK94 pKa = 10.73EE95 pKa = 3.63GDD97 pKa = 3.64VLIDD101 pKa = 2.96VGEE104 pKa = 4.16NVKK107 pKa = 10.47GRR109 pKa = 11.84GDD111 pKa = 3.11KK112 pKa = 11.26GGGFNEE118 pKa = 4.3GANIKK123 pKa = 10.2RR124 pKa = 11.84VVYY127 pKa = 10.31HH128 pKa = 6.54LLEE131 pKa = 4.98GKK133 pKa = 10.08DD134 pKa = 4.06KK135 pKa = 11.01EE136 pKa = 5.41ALLQDD141 pKa = 3.74EE142 pKa = 4.97KK143 pKa = 11.32MLGAYY148 pKa = 7.45MKK150 pKa = 9.3YY151 pKa = 9.92KK152 pKa = 10.52RR153 pKa = 11.84SITEE157 pKa = 3.62MVSEE161 pKa = 4.38VKK163 pKa = 10.4SEE165 pKa = 4.06QEE167 pKa = 4.06TKK169 pKa = 10.44RR170 pKa = 11.84LKK172 pKa = 10.31QDD174 pKa = 3.37LDD176 pKa = 3.69NATLRR181 pKa = 11.84PWQRR185 pKa = 11.84EE186 pKa = 3.8LKK188 pKa = 10.05EE189 pKa = 4.05YY190 pKa = 9.91LTGDD194 pKa = 3.09PHH196 pKa = 6.94DD197 pKa = 4.34RR198 pKa = 11.84HH199 pKa = 7.2IIWYY203 pKa = 9.54IDD205 pKa = 3.35GQGNNGKK212 pKa = 9.11SWFAKK217 pKa = 9.65YY218 pKa = 9.85CVAEE222 pKa = 4.37LGAIRR227 pKa = 11.84LEE229 pKa = 4.2NAKK232 pKa = 8.62TADD235 pKa = 3.42LTHH238 pKa = 7.17AYY240 pKa = 9.89NGEE243 pKa = 3.94RR244 pKa = 11.84VVIFDD249 pKa = 4.88LSRR252 pKa = 11.84TTEE255 pKa = 3.75GHH257 pKa = 5.82FNYY260 pKa = 10.36GALEE264 pKa = 4.28SIKK267 pKa = 10.36NGCVFSSKK275 pKa = 10.46YY276 pKa = 10.42DD277 pKa = 3.4SRR279 pKa = 11.84QKK281 pKa = 9.99MFPIPHH287 pKa = 6.29VIVFANWGPDD297 pKa = 3.06RR298 pKa = 11.84SKK300 pKa = 10.92LSEE303 pKa = 4.33DD304 pKa = 2.84RR305 pKa = 11.84WVTKK309 pKa = 8.48TWSFEE314 pKa = 3.86QEE316 pKa = 4.04TFQTFF321 pKa = 2.92
Molecular weight: 36.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1RL70|A0A0K1RL70_9CIRC Putative capsid protein OS=Palaemonetes sp. common grass shrimp associated circular virus OX=1692259 PE=4 SV=1
MM1 pKa = 7.49LPFAVEE7 pKa = 4.43LAQNAYY13 pKa = 10.06AAYY16 pKa = 9.4EE17 pKa = 4.04LGKK20 pKa = 10.3QGYY23 pKa = 8.85EE24 pKa = 3.99YY25 pKa = 11.08YY26 pKa = 10.43KK27 pKa = 10.58DD28 pKa = 3.61YY29 pKa = 11.38SPYY32 pKa = 9.98VYY34 pKa = 10.29RR35 pKa = 11.84AGRR38 pKa = 11.84GVKK41 pKa = 9.79RR42 pKa = 11.84KK43 pKa = 10.17LGAFLQNEE51 pKa = 4.21QDD53 pKa = 4.33LDD55 pKa = 3.83DD56 pKa = 4.58SPAMRR61 pKa = 11.84RR62 pKa = 11.84MRR64 pKa = 11.84RR65 pKa = 11.84YY66 pKa = 9.46SRR68 pKa = 11.84RR69 pKa = 11.84GMLMRR74 pKa = 11.84RR75 pKa = 11.84RR76 pKa = 11.84FRR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 11.84VFRR85 pKa = 11.84RR86 pKa = 11.84SRR88 pKa = 11.84RR89 pKa = 11.84VFRR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84AFRR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84SGVITSLLRR109 pKa = 11.84FEE111 pKa = 4.81TDD113 pKa = 2.8IKK115 pKa = 11.0LDD117 pKa = 3.88SNGVANNSLYY127 pKa = 11.1FHH129 pKa = 7.04MGTLRR134 pKa = 11.84TKK136 pKa = 10.25INSAYY141 pKa = 9.77IHH143 pKa = 6.33MFDD146 pKa = 3.63EE147 pKa = 5.27FKK149 pKa = 10.62ILSISVVMRR158 pKa = 11.84HH159 pKa = 6.41DD160 pKa = 4.08KK161 pKa = 11.26NMSATDD167 pKa = 3.6NAITEE172 pKa = 4.4LWWTYY177 pKa = 11.61DD178 pKa = 3.23PDD180 pKa = 3.56LTGRR184 pKa = 11.84KK185 pKa = 8.58IDD187 pKa = 3.51MDD189 pKa = 4.3NIKK192 pKa = 9.25MVRR195 pKa = 11.84RR196 pKa = 11.84ARR198 pKa = 11.84SVHH201 pKa = 6.31LGPMGKK207 pKa = 9.38KK208 pKa = 9.51VLKK211 pKa = 10.0CYY213 pKa = 10.16PIFVNPTKK221 pKa = 10.99LEE223 pKa = 4.19GGEE226 pKa = 3.98QQQTISSRR234 pKa = 11.84GSAYY238 pKa = 10.09NRR240 pKa = 11.84PWFDD244 pKa = 3.12AAGLNVGYY252 pKa = 10.32SDD254 pKa = 5.09AMKK257 pKa = 10.23SHH259 pKa = 6.75NGIAWALEE267 pKa = 3.99GQPGRR272 pKa = 11.84GLRR275 pKa = 11.84LDD277 pKa = 3.55YY278 pKa = 10.6AIRR281 pKa = 11.84IAWRR285 pKa = 11.84GRR287 pKa = 11.84RR288 pKa = 11.84DD289 pKa = 3.31GQTYY293 pKa = 9.5TGDD296 pKa = 3.33SS297 pKa = 3.48
MM1 pKa = 7.49LPFAVEE7 pKa = 4.43LAQNAYY13 pKa = 10.06AAYY16 pKa = 9.4EE17 pKa = 4.04LGKK20 pKa = 10.3QGYY23 pKa = 8.85EE24 pKa = 3.99YY25 pKa = 11.08YY26 pKa = 10.43KK27 pKa = 10.58DD28 pKa = 3.61YY29 pKa = 11.38SPYY32 pKa = 9.98VYY34 pKa = 10.29RR35 pKa = 11.84AGRR38 pKa = 11.84GVKK41 pKa = 9.79RR42 pKa = 11.84KK43 pKa = 10.17LGAFLQNEE51 pKa = 4.21QDD53 pKa = 4.33LDD55 pKa = 3.83DD56 pKa = 4.58SPAMRR61 pKa = 11.84RR62 pKa = 11.84MRR64 pKa = 11.84RR65 pKa = 11.84YY66 pKa = 9.46SRR68 pKa = 11.84RR69 pKa = 11.84GMLMRR74 pKa = 11.84RR75 pKa = 11.84RR76 pKa = 11.84FRR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 11.84VFRR85 pKa = 11.84RR86 pKa = 11.84SRR88 pKa = 11.84RR89 pKa = 11.84VFRR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84AFRR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84SGVITSLLRR109 pKa = 11.84FEE111 pKa = 4.81TDD113 pKa = 2.8IKK115 pKa = 11.0LDD117 pKa = 3.88SNGVANNSLYY127 pKa = 11.1FHH129 pKa = 7.04MGTLRR134 pKa = 11.84TKK136 pKa = 10.25INSAYY141 pKa = 9.77IHH143 pKa = 6.33MFDD146 pKa = 3.63EE147 pKa = 5.27FKK149 pKa = 10.62ILSISVVMRR158 pKa = 11.84HH159 pKa = 6.41DD160 pKa = 4.08KK161 pKa = 11.26NMSATDD167 pKa = 3.6NAITEE172 pKa = 4.4LWWTYY177 pKa = 11.61DD178 pKa = 3.23PDD180 pKa = 3.56LTGRR184 pKa = 11.84KK185 pKa = 8.58IDD187 pKa = 3.51MDD189 pKa = 4.3NIKK192 pKa = 9.25MVRR195 pKa = 11.84RR196 pKa = 11.84ARR198 pKa = 11.84SVHH201 pKa = 6.31LGPMGKK207 pKa = 9.38KK208 pKa = 9.51VLKK211 pKa = 10.0CYY213 pKa = 10.16PIFVNPTKK221 pKa = 10.99LEE223 pKa = 4.19GGEE226 pKa = 3.98QQQTISSRR234 pKa = 11.84GSAYY238 pKa = 10.09NRR240 pKa = 11.84PWFDD244 pKa = 3.12AAGLNVGYY252 pKa = 10.32SDD254 pKa = 5.09AMKK257 pKa = 10.23SHH259 pKa = 6.75NGIAWALEE267 pKa = 3.99GQPGRR272 pKa = 11.84GLRR275 pKa = 11.84LDD277 pKa = 3.55YY278 pKa = 10.6AIRR281 pKa = 11.84IAWRR285 pKa = 11.84GRR287 pKa = 11.84RR288 pKa = 11.84DD289 pKa = 3.31GQTYY293 pKa = 9.5TGDD296 pKa = 3.33SS297 pKa = 3.48
Molecular weight: 34.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
618 |
297 |
321 |
309.0 |
35.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.796 ± 0.419 | 1.133 ± 0.546 |
6.149 ± 0.061 | 5.987 ± 1.798 |
4.207 ± 0.114 | 7.929 ± 0.104 |
2.104 ± 0.288 | 4.207 ± 0.348 |
7.443 ± 1.642 | 7.443 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.074 ± 0.894 | 4.531 ± 0.105 |
2.589 ± 0.303 | 3.236 ± 0.141 |
10.032 ± 3.051 | 5.987 ± 0.282 |
5.502 ± 1.003 | 5.016 ± 0.207 |
1.942 ± 0.177 | 4.693 ± 0.708 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
