
Nematocida parisii (strain ERTm3) (Nematode killer fungus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Microsporidia; Microsporidia incertae sedis; Nematocida; Nematocida parisii
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
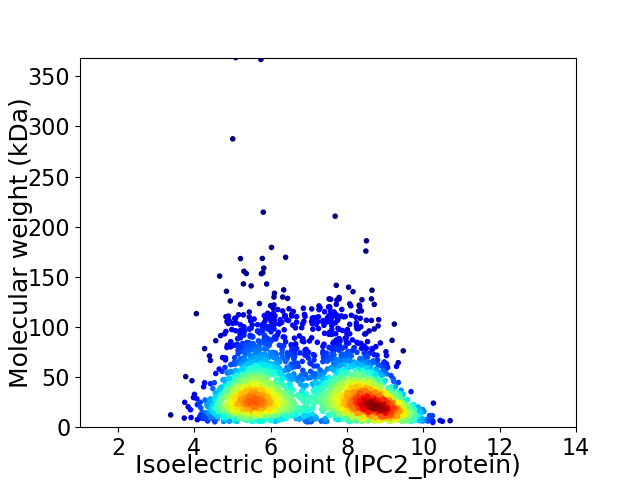
Virtual 2D-PAGE plot for 2724 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
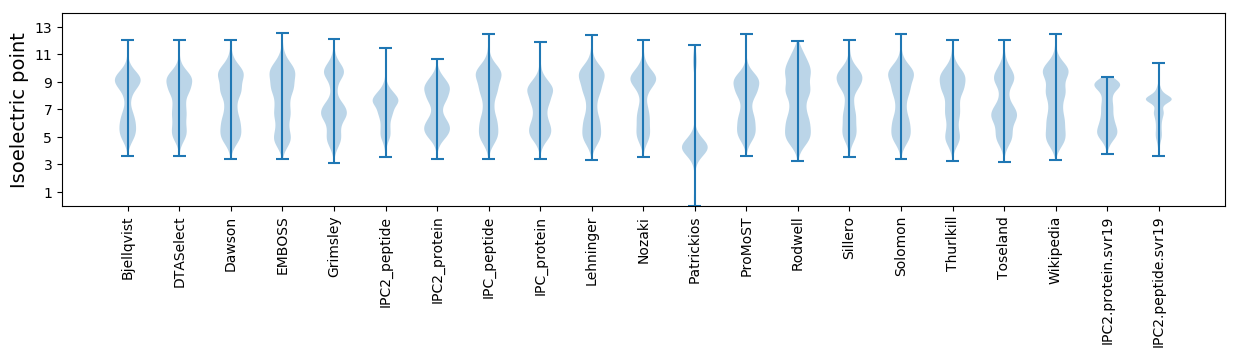
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I3EG79|I3EG79_NEMP3 Uncharacterized protein OS=Nematocida parisii (strain ERTm3) OX=935791 GN=NEQG_01670 PE=4 SV=1
MM1 pKa = 7.6EE2 pKa = 5.43NFMSNYY8 pKa = 9.91DD9 pKa = 3.91EE10 pKa = 4.25QPNTIDD16 pKa = 3.26TPGFEE21 pKa = 4.68TEE23 pKa = 4.33TIVNNDD29 pKa = 3.76EE30 pKa = 3.81LTIIQNIEE38 pKa = 3.8QNVYY42 pKa = 10.54SVSLDD47 pKa = 3.47TAVEE51 pKa = 4.24CGVEE55 pKa = 4.11QIPVDD60 pKa = 3.86PKK62 pKa = 10.4IEE64 pKa = 3.91QLISAFASEE73 pKa = 4.32NRR75 pKa = 11.84FITTNYY81 pKa = 10.12SSNEE85 pKa = 3.54QLIRR89 pKa = 11.84DD90 pKa = 3.98VLSSLEE96 pKa = 4.06KK97 pKa = 10.59KK98 pKa = 10.43GVLFAEE104 pKa = 5.46DD105 pKa = 4.39DD106 pKa = 3.98LSTLSTYY113 pKa = 10.92LNEE116 pKa = 4.17YY117 pKa = 9.69GPLVTDD123 pKa = 3.61VSMSTVLNDD132 pKa = 3.49TNLVTEE138 pKa = 4.86VILTLSEE145 pKa = 3.8NAAEE149 pKa = 3.96ISEE152 pKa = 4.35LNGEE156 pKa = 4.09KK157 pKa = 10.39VLRR160 pKa = 11.84IVSSVDD166 pKa = 3.22LEE168 pKa = 4.29SAYY171 pKa = 11.1SYY173 pKa = 10.92MKK175 pKa = 10.77VCTANQAIVDD185 pKa = 3.87AQSNIFNDD193 pKa = 3.23ICSSLTINMPNLTEE207 pKa = 4.72KK208 pKa = 10.32IVEE211 pKa = 4.24LVEE214 pKa = 4.24TAAEE218 pKa = 3.9PAINAMNN225 pKa = 4.03
MM1 pKa = 7.6EE2 pKa = 5.43NFMSNYY8 pKa = 9.91DD9 pKa = 3.91EE10 pKa = 4.25QPNTIDD16 pKa = 3.26TPGFEE21 pKa = 4.68TEE23 pKa = 4.33TIVNNDD29 pKa = 3.76EE30 pKa = 3.81LTIIQNIEE38 pKa = 3.8QNVYY42 pKa = 10.54SVSLDD47 pKa = 3.47TAVEE51 pKa = 4.24CGVEE55 pKa = 4.11QIPVDD60 pKa = 3.86PKK62 pKa = 10.4IEE64 pKa = 3.91QLISAFASEE73 pKa = 4.32NRR75 pKa = 11.84FITTNYY81 pKa = 10.12SSNEE85 pKa = 3.54QLIRR89 pKa = 11.84DD90 pKa = 3.98VLSSLEE96 pKa = 4.06KK97 pKa = 10.59KK98 pKa = 10.43GVLFAEE104 pKa = 5.46DD105 pKa = 4.39DD106 pKa = 3.98LSTLSTYY113 pKa = 10.92LNEE116 pKa = 4.17YY117 pKa = 9.69GPLVTDD123 pKa = 3.61VSMSTVLNDD132 pKa = 3.49TNLVTEE138 pKa = 4.86VILTLSEE145 pKa = 3.8NAAEE149 pKa = 3.96ISEE152 pKa = 4.35LNGEE156 pKa = 4.09KK157 pKa = 10.39VLRR160 pKa = 11.84IVSSVDD166 pKa = 3.22LEE168 pKa = 4.29SAYY171 pKa = 11.1SYY173 pKa = 10.92MKK175 pKa = 10.77VCTANQAIVDD185 pKa = 3.87AQSNIFNDD193 pKa = 3.23ICSSLTINMPNLTEE207 pKa = 4.72KK208 pKa = 10.32IVEE211 pKa = 4.24LVEE214 pKa = 4.24TAAEE218 pKa = 3.9PAINAMNN225 pKa = 4.03
Molecular weight: 24.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I3ED24|I3ED24_NEMP3 Uncharacterized protein OS=Nematocida parisii (strain ERTm3) OX=935791 GN=NEQG_02690 PE=4 SV=1
MM1 pKa = 7.49VNTQTAIQTAMKK13 pKa = 9.34TVTEE17 pKa = 4.34TATQAINFIRR27 pKa = 11.84EE28 pKa = 4.33SVSEE32 pKa = 4.0ISLDD36 pKa = 4.0GIKK39 pKa = 10.38DD40 pKa = 3.76IIYY43 pKa = 10.48ASTDD47 pKa = 3.22EE48 pKa = 5.29QIDD51 pKa = 3.6LRR53 pKa = 11.84NRR55 pKa = 11.84CILISIAAITLIIVTCVYY73 pKa = 10.44CIIRR77 pKa = 11.84NKK79 pKa = 10.32LKK81 pKa = 10.07AAKK84 pKa = 9.7ARR86 pKa = 11.84RR87 pKa = 11.84QAGQTEE93 pKa = 4.39AEE95 pKa = 4.06RR96 pKa = 11.84QEE98 pKa = 4.18EE99 pKa = 4.47KK100 pKa = 10.66EE101 pKa = 3.82KK102 pKa = 11.11SRR104 pKa = 11.84CKK106 pKa = 9.6KK107 pKa = 9.28TSTSKK112 pKa = 9.97CSRR115 pKa = 11.84YY116 pKa = 9.34NRR118 pKa = 11.84RR119 pKa = 11.84STYY122 pKa = 7.86TTDD125 pKa = 3.0INRR128 pKa = 11.84STSTGCQASTSRR140 pKa = 11.84RR141 pKa = 11.84RR142 pKa = 11.84KK143 pKa = 7.83NTSRR147 pKa = 11.84CKK149 pKa = 9.59KK150 pKa = 9.24TSTCKK155 pKa = 10.23CSRR158 pKa = 11.84KK159 pKa = 8.53KK160 pKa = 9.31TSRR163 pKa = 11.84CNKK166 pKa = 6.83KK167 pKa = 7.31TRR169 pKa = 11.84RR170 pKa = 11.84RR171 pKa = 11.84KK172 pKa = 9.8KK173 pKa = 7.99NTSRR177 pKa = 11.84RR178 pKa = 11.84SKK180 pKa = 10.68KK181 pKa = 8.37KK182 pKa = 8.36TRR184 pKa = 11.84RR185 pKa = 11.84CRR187 pKa = 3.3
MM1 pKa = 7.49VNTQTAIQTAMKK13 pKa = 9.34TVTEE17 pKa = 4.34TATQAINFIRR27 pKa = 11.84EE28 pKa = 4.33SVSEE32 pKa = 4.0ISLDD36 pKa = 4.0GIKK39 pKa = 10.38DD40 pKa = 3.76IIYY43 pKa = 10.48ASTDD47 pKa = 3.22EE48 pKa = 5.29QIDD51 pKa = 3.6LRR53 pKa = 11.84NRR55 pKa = 11.84CILISIAAITLIIVTCVYY73 pKa = 10.44CIIRR77 pKa = 11.84NKK79 pKa = 10.32LKK81 pKa = 10.07AAKK84 pKa = 9.7ARR86 pKa = 11.84RR87 pKa = 11.84QAGQTEE93 pKa = 4.39AEE95 pKa = 4.06RR96 pKa = 11.84QEE98 pKa = 4.18EE99 pKa = 4.47KK100 pKa = 10.66EE101 pKa = 3.82KK102 pKa = 11.11SRR104 pKa = 11.84CKK106 pKa = 9.6KK107 pKa = 9.28TSTSKK112 pKa = 9.97CSRR115 pKa = 11.84YY116 pKa = 9.34NRR118 pKa = 11.84RR119 pKa = 11.84STYY122 pKa = 7.86TTDD125 pKa = 3.0INRR128 pKa = 11.84STSTGCQASTSRR140 pKa = 11.84RR141 pKa = 11.84RR142 pKa = 11.84KK143 pKa = 7.83NTSRR147 pKa = 11.84CKK149 pKa = 9.59KK150 pKa = 9.24TSTCKK155 pKa = 10.23CSRR158 pKa = 11.84KK159 pKa = 8.53KK160 pKa = 9.31TSRR163 pKa = 11.84CNKK166 pKa = 6.83KK167 pKa = 7.31TRR169 pKa = 11.84RR170 pKa = 11.84RR171 pKa = 11.84KK172 pKa = 9.8KK173 pKa = 7.99NTSRR177 pKa = 11.84RR178 pKa = 11.84SKK180 pKa = 10.68KK181 pKa = 8.37KK182 pKa = 8.36TRR184 pKa = 11.84RR185 pKa = 11.84CRR187 pKa = 3.3
Molecular weight: 21.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
955631 |
41 |
3274 |
350.8 |
40.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.85 ± 0.047 | 1.772 ± 0.021 |
4.74 ± 0.032 | 7.532 ± 0.059 |
4.036 ± 0.035 | 4.393 ± 0.042 |
2.042 ± 0.02 | 9.315 ± 0.052 |
8.384 ± 0.048 | 9.182 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.605 ± 0.02 | 5.952 ± 0.058 |
3.375 ± 0.037 | 3.066 ± 0.028 |
4.214 ± 0.033 | 8.238 ± 0.059 |
5.675 ± 0.032 | 5.641 ± 0.036 |
0.665 ± 0.01 | 4.323 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
