
Pteropus associated gemycircularvirus 8
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus ptero8
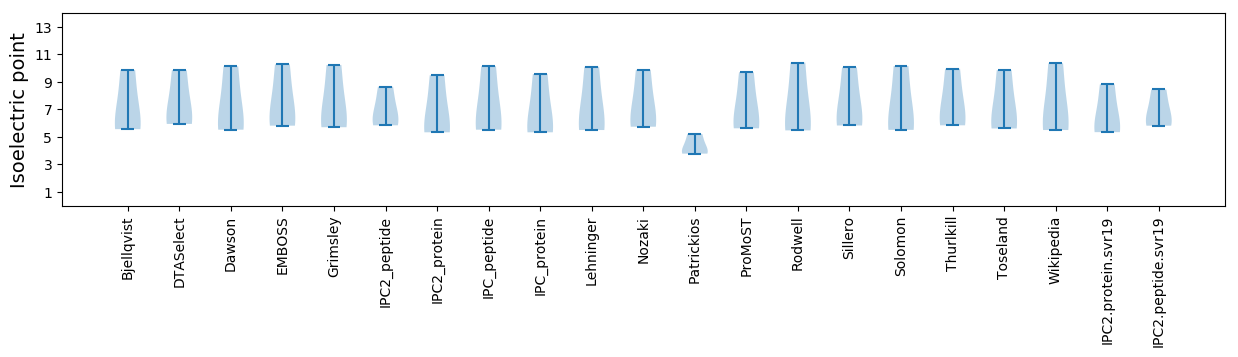
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
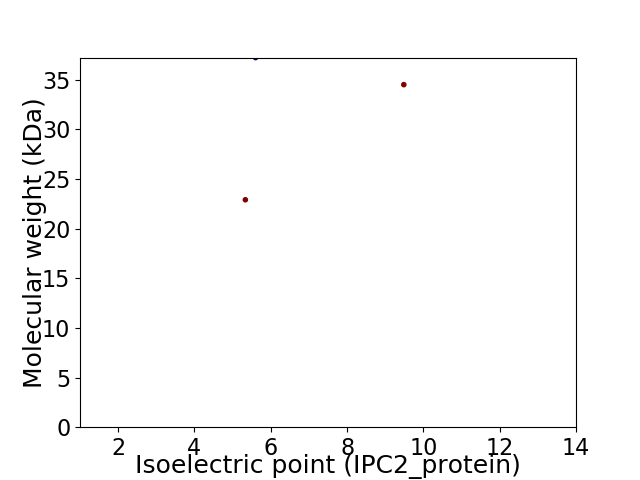
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140CTQ2|A0A140CTQ2_9VIRU RepA OS=Pteropus associated gemycircularvirus 8 OX=1985402 PE=3 SV=1
MM1 pKa = 7.72APFILKK7 pKa = 9.72NVRR10 pKa = 11.84RR11 pKa = 11.84ALLTYY16 pKa = 7.65SQCGTLDD23 pKa = 3.44PFAVSDD29 pKa = 3.63HH30 pKa = 6.2FSNLGAEE37 pKa = 4.53CIVGRR42 pKa = 11.84EE43 pKa = 3.75RR44 pKa = 11.84HH45 pKa = 5.94ADD47 pKa = 3.43GGLHH51 pKa = 5.93LHH53 pKa = 6.03VFVDD57 pKa = 4.61FGRR60 pKa = 11.84QFSSRR65 pKa = 11.84KK66 pKa = 7.81TDD68 pKa = 3.11VFDD71 pKa = 4.59VGGHH75 pKa = 6.1HH76 pKa = 7.25PNIAKK81 pKa = 10.36CGRR84 pKa = 11.84TPWKK88 pKa = 10.45AYY90 pKa = 10.52DD91 pKa = 3.69YY92 pKa = 10.6AIKK95 pKa = 10.92DD96 pKa = 3.52GDD98 pKa = 4.09VVAGGLDD105 pKa = 3.48RR106 pKa = 11.84PIEE109 pKa = 4.01EE110 pKa = 4.85SGDD113 pKa = 3.54GSGTIDD119 pKa = 3.9STWHH123 pKa = 6.33SILAAEE129 pKa = 4.46TPEE132 pKa = 4.2EE133 pKa = 4.41FWALCHH139 pKa = 7.01DD140 pKa = 4.97LAPRR144 pKa = 11.84DD145 pKa = 3.72LARR148 pKa = 11.84SFPSLQKK155 pKa = 10.22YY156 pKa = 9.88CDD158 pKa = 2.84WRR160 pKa = 11.84YY161 pKa = 10.12RR162 pKa = 11.84PVTRR166 pKa = 11.84PYY168 pKa = 8.77CTPDD172 pKa = 3.0GYY174 pKa = 11.41EE175 pKa = 4.13FDD177 pKa = 3.55TSRR180 pKa = 11.84APEE183 pKa = 4.1LNEE186 pKa = 3.56WLLQANLGLRR196 pKa = 11.84GNWDD200 pKa = 2.89QGMCC204 pKa = 4.51
MM1 pKa = 7.72APFILKK7 pKa = 9.72NVRR10 pKa = 11.84RR11 pKa = 11.84ALLTYY16 pKa = 7.65SQCGTLDD23 pKa = 3.44PFAVSDD29 pKa = 3.63HH30 pKa = 6.2FSNLGAEE37 pKa = 4.53CIVGRR42 pKa = 11.84EE43 pKa = 3.75RR44 pKa = 11.84HH45 pKa = 5.94ADD47 pKa = 3.43GGLHH51 pKa = 5.93LHH53 pKa = 6.03VFVDD57 pKa = 4.61FGRR60 pKa = 11.84QFSSRR65 pKa = 11.84KK66 pKa = 7.81TDD68 pKa = 3.11VFDD71 pKa = 4.59VGGHH75 pKa = 6.1HH76 pKa = 7.25PNIAKK81 pKa = 10.36CGRR84 pKa = 11.84TPWKK88 pKa = 10.45AYY90 pKa = 10.52DD91 pKa = 3.69YY92 pKa = 10.6AIKK95 pKa = 10.92DD96 pKa = 3.52GDD98 pKa = 4.09VVAGGLDD105 pKa = 3.48RR106 pKa = 11.84PIEE109 pKa = 4.01EE110 pKa = 4.85SGDD113 pKa = 3.54GSGTIDD119 pKa = 3.9STWHH123 pKa = 6.33SILAAEE129 pKa = 4.46TPEE132 pKa = 4.2EE133 pKa = 4.41FWALCHH139 pKa = 7.01DD140 pKa = 4.97LAPRR144 pKa = 11.84DD145 pKa = 3.72LARR148 pKa = 11.84SFPSLQKK155 pKa = 10.22YY156 pKa = 9.88CDD158 pKa = 2.84WRR160 pKa = 11.84YY161 pKa = 10.12RR162 pKa = 11.84PVTRR166 pKa = 11.84PYY168 pKa = 8.77CTPDD172 pKa = 3.0GYY174 pKa = 11.41EE175 pKa = 4.13FDD177 pKa = 3.55TSRR180 pKa = 11.84APEE183 pKa = 4.1LNEE186 pKa = 3.56WLLQANLGLRR196 pKa = 11.84GNWDD200 pKa = 2.89QGMCC204 pKa = 4.51
Molecular weight: 22.89 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTQ1|A0A140CTQ1_9VIRU Replication-associated protein OS=Pteropus associated gemycircularvirus 8 OX=1985402 PE=3 SV=1
MM1 pKa = 7.48PRR3 pKa = 11.84YY4 pKa = 7.13ATRR7 pKa = 11.84RR8 pKa = 11.84ISGRR12 pKa = 11.84YY13 pKa = 9.36SKK15 pKa = 9.62TKK17 pKa = 8.85QANRR21 pKa = 11.84YY22 pKa = 6.18KK23 pKa = 10.74AKK25 pKa = 10.62YY26 pKa = 7.84KK27 pKa = 10.37RR28 pKa = 11.84KK29 pKa = 8.77YY30 pKa = 8.04KK31 pKa = 10.68AKK33 pKa = 9.6RR34 pKa = 11.84KK35 pKa = 8.95YY36 pKa = 11.0GGMSKK41 pKa = 10.56KK42 pKa = 10.48KK43 pKa = 10.28ILDD46 pKa = 3.1LTAIKK51 pKa = 10.42KK52 pKa = 9.71RR53 pKa = 11.84DD54 pKa = 3.51AMLVSTNITLASPSGSTSFNNNGAVLGPNRR84 pKa = 11.84TYY86 pKa = 10.53FIPWIATWRR95 pKa = 11.84DD96 pKa = 2.86NSTVSGGAQTGTRR109 pKa = 11.84FDD111 pKa = 3.59TATRR115 pKa = 11.84TATTCYY121 pKa = 9.61MRR123 pKa = 11.84GLKK126 pKa = 9.11EE127 pKa = 3.71QIYY130 pKa = 9.44FIQNDD135 pKa = 3.66GSQWMWRR142 pKa = 11.84RR143 pKa = 11.84ICFKK147 pKa = 10.04YY148 pKa = 10.16RR149 pKa = 11.84GPVVYY154 pKa = 9.86QQSKK158 pKa = 10.15AGYY161 pKa = 7.42TLAVEE166 pKa = 4.66TSNGFQRR173 pKa = 11.84LYY175 pKa = 11.62NNINDD180 pKa = 3.75SSGANMLNTMIGIMFRR196 pKa = 11.84GQVGVDD202 pKa = 2.95YY203 pKa = 11.39GNFFNAPTDD212 pKa = 3.49NTRR215 pKa = 11.84CEE217 pKa = 3.68IVYY220 pKa = 10.73DD221 pKa = 3.84KK222 pKa = 10.86TRR224 pKa = 11.84SLATGNQNGYY234 pKa = 7.54SRR236 pKa = 11.84RR237 pKa = 11.84VSLWHH242 pKa = 6.47GMNKK246 pKa = 9.65NLVYY250 pKa = 10.85DD251 pKa = 4.21DD252 pKa = 5.5DD253 pKa = 4.32EE254 pKa = 6.95AGGAIAAGNASVSDD268 pKa = 3.49KK269 pKa = 11.15RR270 pKa = 11.84GMGDD274 pKa = 3.56YY275 pKa = 10.64YY276 pKa = 11.16VVDD279 pKa = 3.65MFICEE284 pKa = 4.31APQSSSSQLIVVPEE298 pKa = 3.81ATLYY302 pKa = 9.06WHH304 pKa = 7.08EE305 pKa = 4.26KK306 pKa = 9.3
MM1 pKa = 7.48PRR3 pKa = 11.84YY4 pKa = 7.13ATRR7 pKa = 11.84RR8 pKa = 11.84ISGRR12 pKa = 11.84YY13 pKa = 9.36SKK15 pKa = 9.62TKK17 pKa = 8.85QANRR21 pKa = 11.84YY22 pKa = 6.18KK23 pKa = 10.74AKK25 pKa = 10.62YY26 pKa = 7.84KK27 pKa = 10.37RR28 pKa = 11.84KK29 pKa = 8.77YY30 pKa = 8.04KK31 pKa = 10.68AKK33 pKa = 9.6RR34 pKa = 11.84KK35 pKa = 8.95YY36 pKa = 11.0GGMSKK41 pKa = 10.56KK42 pKa = 10.48KK43 pKa = 10.28ILDD46 pKa = 3.1LTAIKK51 pKa = 10.42KK52 pKa = 9.71RR53 pKa = 11.84DD54 pKa = 3.51AMLVSTNITLASPSGSTSFNNNGAVLGPNRR84 pKa = 11.84TYY86 pKa = 10.53FIPWIATWRR95 pKa = 11.84DD96 pKa = 2.86NSTVSGGAQTGTRR109 pKa = 11.84FDD111 pKa = 3.59TATRR115 pKa = 11.84TATTCYY121 pKa = 9.61MRR123 pKa = 11.84GLKK126 pKa = 9.11EE127 pKa = 3.71QIYY130 pKa = 9.44FIQNDD135 pKa = 3.66GSQWMWRR142 pKa = 11.84RR143 pKa = 11.84ICFKK147 pKa = 10.04YY148 pKa = 10.16RR149 pKa = 11.84GPVVYY154 pKa = 9.86QQSKK158 pKa = 10.15AGYY161 pKa = 7.42TLAVEE166 pKa = 4.66TSNGFQRR173 pKa = 11.84LYY175 pKa = 11.62NNINDD180 pKa = 3.75SSGANMLNTMIGIMFRR196 pKa = 11.84GQVGVDD202 pKa = 2.95YY203 pKa = 11.39GNFFNAPTDD212 pKa = 3.49NTRR215 pKa = 11.84CEE217 pKa = 3.68IVYY220 pKa = 10.73DD221 pKa = 3.84KK222 pKa = 10.86TRR224 pKa = 11.84SLATGNQNGYY234 pKa = 7.54SRR236 pKa = 11.84RR237 pKa = 11.84VSLWHH242 pKa = 6.47GMNKK246 pKa = 9.65NLVYY250 pKa = 10.85DD251 pKa = 4.21DD252 pKa = 5.5DD253 pKa = 4.32EE254 pKa = 6.95AGGAIAAGNASVSDD268 pKa = 3.49KK269 pKa = 11.15RR270 pKa = 11.84GMGDD274 pKa = 3.56YY275 pKa = 10.64YY276 pKa = 11.16VVDD279 pKa = 3.65MFICEE284 pKa = 4.31APQSSSSQLIVVPEE298 pKa = 3.81ATLYY302 pKa = 9.06WHH304 pKa = 7.08EE305 pKa = 4.26KK306 pKa = 9.3
Molecular weight: 34.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
842 |
204 |
332 |
280.7 |
31.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.838 ± 0.003 | 2.257 ± 0.502 |
7.126 ± 1.017 | 3.682 ± 0.655 |
4.513 ± 0.551 | 9.145 ± 0.142 |
2.613 ± 0.88 | 4.513 ± 0.421 |
4.869 ± 0.961 | 7.363 ± 1.233 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.781 ± 0.799 | 4.513 ± 1.19 |
4.632 ± 0.9 | 3.088 ± 0.381 |
6.888 ± 0.382 | 6.888 ± 0.473 |
5.819 ± 0.892 | 5.107 ± 0.077 |
2.732 ± 0.342 | 4.632 ± 0.851 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
