
Picobirnavirus sp.
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Picobirnaviridae; Picobirnavirus; unclassified Picobirnavirus
Average proteome isoelectric point is 7.03
Get precalculated fractions of proteins
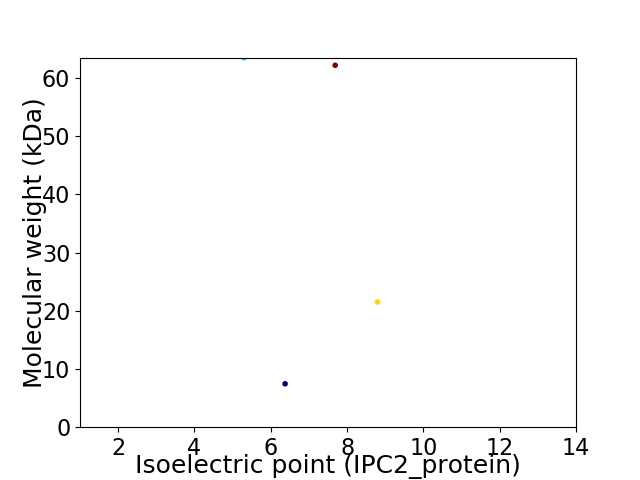
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A291S0D7|A0A291S0D7_9VIRU Uncharacterized protein OS=Picobirnavirus sp. OX=1907787 PE=4 SV=1
MM1 pKa = 7.21NKK3 pKa = 8.34KK4 pKa = 8.56TGRR7 pKa = 11.84NQIPKK12 pKa = 9.74SNKK15 pKa = 9.33AKK17 pKa = 10.72ASTTGRR23 pKa = 11.84KK24 pKa = 8.66GRR26 pKa = 11.84GMRR29 pKa = 11.84DD30 pKa = 3.15QHH32 pKa = 6.1SQRR35 pKa = 11.84SDD37 pKa = 2.93NRR39 pKa = 11.84NVNSSGNYY47 pKa = 9.84NGDD50 pKa = 2.8IDD52 pKa = 5.7APATNNPDD60 pKa = 2.93WWNKK64 pKa = 9.19SATFADD70 pKa = 4.2SVRR73 pKa = 11.84IPFNRR78 pKa = 11.84ITGYY82 pKa = 9.86PISKK86 pKa = 10.23NDD88 pKa = 3.85LLPRR92 pKa = 11.84SIFQDD97 pKa = 3.28TTIVADD103 pKa = 3.85EE104 pKa = 4.86SYY106 pKa = 10.43PEE108 pKa = 5.2AIAIPNLMCIHH119 pKa = 6.82YY120 pKa = 10.36IPFTGACSNWNDD132 pKa = 4.38PINRR136 pKa = 11.84SYY138 pKa = 10.89MSVYY142 pKa = 10.37SALAAKK148 pKa = 9.68TSGAMQINQPDD159 pKa = 3.56LAMTTEE165 pKa = 4.28AANSVIVLIYY175 pKa = 9.37EE176 pKa = 4.55LKK178 pKa = 10.3RR179 pKa = 11.84ALGIANLYY187 pKa = 8.79SQQNKK192 pKa = 7.22TLPYY196 pKa = 9.78HH197 pKa = 6.32VLQALHH203 pKa = 7.21IDD205 pKa = 3.57PDD207 pKa = 4.68SIIGHH212 pKa = 5.31QDD214 pKa = 3.1EE215 pKa = 4.5IRR217 pKa = 11.84RR218 pKa = 11.84EE219 pKa = 3.76LNYY222 pKa = 11.0YY223 pKa = 9.91IGALNSLALPAYY235 pKa = 8.68TDD237 pKa = 2.74IHH239 pKa = 6.48ARR241 pKa = 11.84RR242 pKa = 11.84AQLAGNVYY250 pKa = 10.79ADD252 pKa = 3.96EE253 pKa = 5.39DD254 pKa = 3.93DD255 pKa = 3.77VRR257 pKa = 11.84AAFYY261 pKa = 10.39AYY263 pKa = 8.47VTAGYY268 pKa = 9.98YY269 pKa = 10.31EE270 pKa = 4.43YY271 pKa = 11.27VDD273 pKa = 3.72TADD276 pKa = 5.11DD277 pKa = 3.71ISDD280 pKa = 3.79VEE282 pKa = 4.24HH283 pKa = 6.84LKK285 pKa = 10.34FVYY288 pKa = 9.2MAASTPRR295 pKa = 11.84TADD298 pKa = 2.98SYY300 pKa = 10.69LTAIGDD306 pKa = 4.34CINAIRR312 pKa = 11.84NSSAFGLISAAVQRR326 pKa = 11.84GFGDD330 pKa = 3.61RR331 pKa = 11.84NLLTVEE337 pKa = 4.14MVPVDD342 pKa = 4.05FVVIPAVDD350 pKa = 3.34RR351 pKa = 11.84YY352 pKa = 7.66MTYY355 pKa = 8.53QTQNADD361 pKa = 4.21FIGHH365 pKa = 6.1YY366 pKa = 10.38NDD368 pKa = 5.16LDD370 pKa = 4.02SKK372 pKa = 11.04SLHH375 pKa = 5.87IVQNPVRR382 pKa = 11.84NALEE386 pKa = 4.16VIPTIKK392 pKa = 10.4SNSSHH397 pKa = 6.56YY398 pKa = 9.59PEE400 pKa = 4.11NWACIVNDD408 pKa = 3.85NKK410 pKa = 10.27WLNSYY415 pKa = 11.04DD416 pKa = 4.22GITDD420 pKa = 3.48ADD422 pKa = 3.95FVIEE426 pKa = 4.01ATRR429 pKa = 11.84LKK431 pKa = 10.84VATTAIEE438 pKa = 4.4DD439 pKa = 3.64PGFEE443 pKa = 4.15YY444 pKa = 10.98GMDD447 pKa = 3.24IPLDD451 pKa = 3.59VTGTEE456 pKa = 3.57ICLNFHH462 pKa = 5.42IWKK465 pKa = 10.18VEE467 pKa = 3.62IVNGKK472 pKa = 7.28EE473 pKa = 3.76TLTATIHH480 pKa = 6.46KK481 pKa = 8.97SWNYY485 pKa = 8.89VANADD490 pKa = 3.99PVAMATQVKK499 pKa = 8.98FMCNLSKK506 pKa = 10.77FKK508 pKa = 10.66NHH510 pKa = 6.53PLINFILKK518 pKa = 9.24TDD520 pKa = 3.99DD521 pKa = 3.5VVKK524 pKa = 10.03TGVITPIPFGDD535 pKa = 4.25LYY537 pKa = 10.71RR538 pKa = 11.84YY539 pKa = 8.62TIMSGSALQGIHH551 pKa = 5.21EE552 pKa = 4.61TALQSIYY559 pKa = 10.81TLVPSLDD566 pKa = 3.37IVGEE570 pKa = 4.17KK571 pKa = 10.38
MM1 pKa = 7.21NKK3 pKa = 8.34KK4 pKa = 8.56TGRR7 pKa = 11.84NQIPKK12 pKa = 9.74SNKK15 pKa = 9.33AKK17 pKa = 10.72ASTTGRR23 pKa = 11.84KK24 pKa = 8.66GRR26 pKa = 11.84GMRR29 pKa = 11.84DD30 pKa = 3.15QHH32 pKa = 6.1SQRR35 pKa = 11.84SDD37 pKa = 2.93NRR39 pKa = 11.84NVNSSGNYY47 pKa = 9.84NGDD50 pKa = 2.8IDD52 pKa = 5.7APATNNPDD60 pKa = 2.93WWNKK64 pKa = 9.19SATFADD70 pKa = 4.2SVRR73 pKa = 11.84IPFNRR78 pKa = 11.84ITGYY82 pKa = 9.86PISKK86 pKa = 10.23NDD88 pKa = 3.85LLPRR92 pKa = 11.84SIFQDD97 pKa = 3.28TTIVADD103 pKa = 3.85EE104 pKa = 4.86SYY106 pKa = 10.43PEE108 pKa = 5.2AIAIPNLMCIHH119 pKa = 6.82YY120 pKa = 10.36IPFTGACSNWNDD132 pKa = 4.38PINRR136 pKa = 11.84SYY138 pKa = 10.89MSVYY142 pKa = 10.37SALAAKK148 pKa = 9.68TSGAMQINQPDD159 pKa = 3.56LAMTTEE165 pKa = 4.28AANSVIVLIYY175 pKa = 9.37EE176 pKa = 4.55LKK178 pKa = 10.3RR179 pKa = 11.84ALGIANLYY187 pKa = 8.79SQQNKK192 pKa = 7.22TLPYY196 pKa = 9.78HH197 pKa = 6.32VLQALHH203 pKa = 7.21IDD205 pKa = 3.57PDD207 pKa = 4.68SIIGHH212 pKa = 5.31QDD214 pKa = 3.1EE215 pKa = 4.5IRR217 pKa = 11.84RR218 pKa = 11.84EE219 pKa = 3.76LNYY222 pKa = 11.0YY223 pKa = 9.91IGALNSLALPAYY235 pKa = 8.68TDD237 pKa = 2.74IHH239 pKa = 6.48ARR241 pKa = 11.84RR242 pKa = 11.84AQLAGNVYY250 pKa = 10.79ADD252 pKa = 3.96EE253 pKa = 5.39DD254 pKa = 3.93DD255 pKa = 3.77VRR257 pKa = 11.84AAFYY261 pKa = 10.39AYY263 pKa = 8.47VTAGYY268 pKa = 9.98YY269 pKa = 10.31EE270 pKa = 4.43YY271 pKa = 11.27VDD273 pKa = 3.72TADD276 pKa = 5.11DD277 pKa = 3.71ISDD280 pKa = 3.79VEE282 pKa = 4.24HH283 pKa = 6.84LKK285 pKa = 10.34FVYY288 pKa = 9.2MAASTPRR295 pKa = 11.84TADD298 pKa = 2.98SYY300 pKa = 10.69LTAIGDD306 pKa = 4.34CINAIRR312 pKa = 11.84NSSAFGLISAAVQRR326 pKa = 11.84GFGDD330 pKa = 3.61RR331 pKa = 11.84NLLTVEE337 pKa = 4.14MVPVDD342 pKa = 4.05FVVIPAVDD350 pKa = 3.34RR351 pKa = 11.84YY352 pKa = 7.66MTYY355 pKa = 8.53QTQNADD361 pKa = 4.21FIGHH365 pKa = 6.1YY366 pKa = 10.38NDD368 pKa = 5.16LDD370 pKa = 4.02SKK372 pKa = 11.04SLHH375 pKa = 5.87IVQNPVRR382 pKa = 11.84NALEE386 pKa = 4.16VIPTIKK392 pKa = 10.4SNSSHH397 pKa = 6.56YY398 pKa = 9.59PEE400 pKa = 4.11NWACIVNDD408 pKa = 3.85NKK410 pKa = 10.27WLNSYY415 pKa = 11.04DD416 pKa = 4.22GITDD420 pKa = 3.48ADD422 pKa = 3.95FVIEE426 pKa = 4.01ATRR429 pKa = 11.84LKK431 pKa = 10.84VATTAIEE438 pKa = 4.4DD439 pKa = 3.64PGFEE443 pKa = 4.15YY444 pKa = 10.98GMDD447 pKa = 3.24IPLDD451 pKa = 3.59VTGTEE456 pKa = 3.57ICLNFHH462 pKa = 5.42IWKK465 pKa = 10.18VEE467 pKa = 3.62IVNGKK472 pKa = 7.28EE473 pKa = 3.76TLTATIHH480 pKa = 6.46KK481 pKa = 8.97SWNYY485 pKa = 8.89VANADD490 pKa = 3.99PVAMATQVKK499 pKa = 8.98FMCNLSKK506 pKa = 10.77FKK508 pKa = 10.66NHH510 pKa = 6.53PLINFILKK518 pKa = 9.24TDD520 pKa = 3.99DD521 pKa = 3.5VVKK524 pKa = 10.03TGVITPIPFGDD535 pKa = 4.25LYY537 pKa = 10.71RR538 pKa = 11.84YY539 pKa = 8.62TIMSGSALQGIHH551 pKa = 5.21EE552 pKa = 4.61TALQSIYY559 pKa = 10.81TLVPSLDD566 pKa = 3.37IVGEE570 pKa = 4.17KK571 pKa = 10.38
Molecular weight: 63.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A291S0E7|A0A291S0E7_9VIRU Uncharacterized protein OS=Picobirnavirus sp. OX=1907787 PE=4 SV=1
MM1 pKa = 7.47KK2 pKa = 10.11GGHH5 pKa = 5.91SFMTTQAIEE14 pKa = 3.96YY15 pKa = 9.04LKK17 pKa = 10.81LRR19 pKa = 11.84EE20 pKa = 4.15TEE22 pKa = 3.93RR23 pKa = 11.84SNKK26 pKa = 9.29ARR28 pKa = 11.84EE29 pKa = 4.28KK30 pKa = 8.47EE31 pKa = 4.11THH33 pKa = 6.3RR34 pKa = 11.84ANLAQEE40 pKa = 4.77NISTANLAEE49 pKa = 4.28TTRR52 pKa = 11.84SNKK55 pKa = 9.96AKK57 pKa = 10.45EE58 pKa = 4.35SISLDD63 pKa = 3.19TLAEE67 pKa = 4.31TKK69 pKa = 10.34RR70 pKa = 11.84SNQAKK75 pKa = 9.63EE76 pKa = 4.1AIEE79 pKa = 3.91TSKK82 pKa = 10.53FAEE85 pKa = 4.64TIRR88 pKa = 11.84SNLAKK93 pKa = 10.27EE94 pKa = 4.54FEE96 pKa = 4.52TQRR99 pKa = 11.84SNLAKK104 pKa = 9.72EE105 pKa = 4.26QEE107 pKa = 4.49TKK109 pKa = 10.47RR110 pKa = 11.84SNIAKK115 pKa = 9.18EE116 pKa = 3.83QEE118 pKa = 4.35TYY120 pKa = 10.34RR121 pKa = 11.84SNVARR126 pKa = 11.84EE127 pKa = 3.8RR128 pKa = 11.84EE129 pKa = 4.35TYY131 pKa = 9.88RR132 pKa = 11.84SNVAQEE138 pKa = 3.85SNTRR142 pKa = 11.84YY143 pKa = 8.15KK144 pKa = 10.89TNLDD148 pKa = 3.59NIGYY152 pKa = 9.48ISQAAALDD160 pKa = 3.79DD161 pKa = 3.8MGYY164 pKa = 10.4NPAKK168 pKa = 10.51SSLLMLYY175 pKa = 10.17DD176 pKa = 3.94AARR179 pKa = 11.84GLVPKK184 pKa = 10.6LSVSFKK190 pKa = 10.97
MM1 pKa = 7.47KK2 pKa = 10.11GGHH5 pKa = 5.91SFMTTQAIEE14 pKa = 3.96YY15 pKa = 9.04LKK17 pKa = 10.81LRR19 pKa = 11.84EE20 pKa = 4.15TEE22 pKa = 3.93RR23 pKa = 11.84SNKK26 pKa = 9.29ARR28 pKa = 11.84EE29 pKa = 4.28KK30 pKa = 8.47EE31 pKa = 4.11THH33 pKa = 6.3RR34 pKa = 11.84ANLAQEE40 pKa = 4.77NISTANLAEE49 pKa = 4.28TTRR52 pKa = 11.84SNKK55 pKa = 9.96AKK57 pKa = 10.45EE58 pKa = 4.35SISLDD63 pKa = 3.19TLAEE67 pKa = 4.31TKK69 pKa = 10.34RR70 pKa = 11.84SNQAKK75 pKa = 9.63EE76 pKa = 4.1AIEE79 pKa = 3.91TSKK82 pKa = 10.53FAEE85 pKa = 4.64TIRR88 pKa = 11.84SNLAKK93 pKa = 10.27EE94 pKa = 4.54FEE96 pKa = 4.52TQRR99 pKa = 11.84SNLAKK104 pKa = 9.72EE105 pKa = 4.26QEE107 pKa = 4.49TKK109 pKa = 10.47RR110 pKa = 11.84SNIAKK115 pKa = 9.18EE116 pKa = 3.83QEE118 pKa = 4.35TYY120 pKa = 10.34RR121 pKa = 11.84SNVARR126 pKa = 11.84EE127 pKa = 3.8RR128 pKa = 11.84EE129 pKa = 4.35TYY131 pKa = 9.88RR132 pKa = 11.84SNVAQEE138 pKa = 3.85SNTRR142 pKa = 11.84YY143 pKa = 8.15KK144 pKa = 10.89TNLDD148 pKa = 3.59NIGYY152 pKa = 9.48ISQAAALDD160 pKa = 3.79DD161 pKa = 3.8MGYY164 pKa = 10.4NPAKK168 pKa = 10.51SSLLMLYY175 pKa = 10.17DD176 pKa = 3.94AARR179 pKa = 11.84GLVPKK184 pKa = 10.6LSVSFKK190 pKa = 10.97
Molecular weight: 21.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1366 |
63 |
571 |
341.5 |
38.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.053 ± 1.422 | 1.098 ± 0.237 |
6.369 ± 0.969 | 5.124 ± 1.182 |
3.221 ± 0.424 | 5.198 ± 0.688 |
2.123 ± 0.561 | 6.735 ± 1.199 |
5.637 ± 0.922 | 7.613 ± 0.541 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.562 ± 0.298 | 6.369 ± 0.853 |
4.026 ± 0.574 | 3.807 ± 0.42 |
5.71 ± 0.832 | 7.394 ± 0.536 |
6.369 ± 0.714 | 6.003 ± 0.733 |
1.537 ± 0.531 | 5.051 ± 0.28 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
