
Indian encephalitis associated cyclovirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Cyclovirus; Human associated cyclovirus 12
Average proteome isoelectric point is 8.68
Get precalculated fractions of proteins
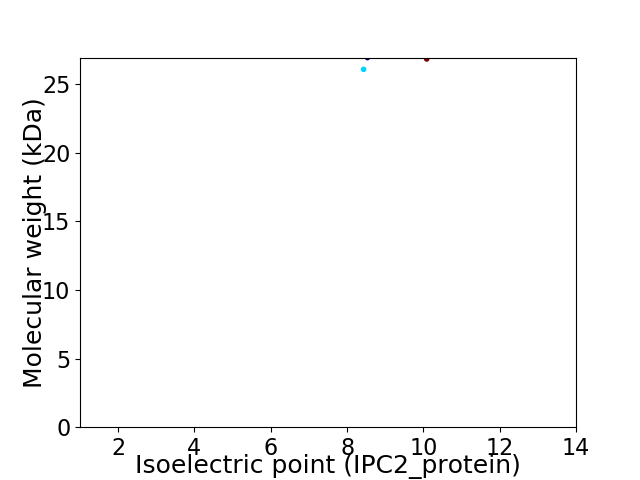
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
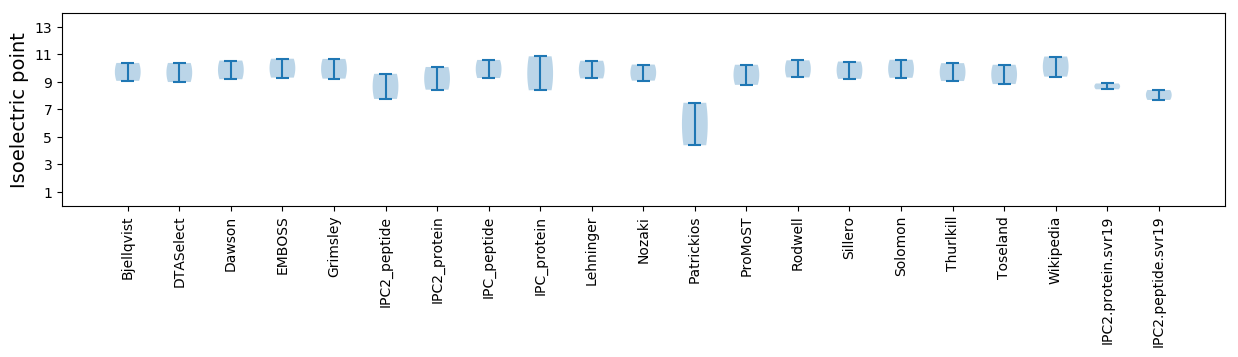
Protein with the lowest isoelectric point:
>tr|A0A1N7TEH5|A0A1N7TEH5_9CIRC Replication-associated protein OS=Indian encephalitis associated cyclovirus OX=1755290 PE=4 SV=1
MM1 pKa = 7.39EE2 pKa = 5.14RR3 pKa = 11.84VTLKK7 pKa = 10.63RR8 pKa = 11.84FCFTLNNYY16 pKa = 10.45DD17 pKa = 3.62EE18 pKa = 5.32DD19 pKa = 3.74RR20 pKa = 11.84VKK22 pKa = 10.42ILKK25 pKa = 10.11EE26 pKa = 3.99FMSTKK31 pKa = 9.84CKK33 pKa = 8.81YY34 pKa = 10.62AIMGFEE40 pKa = 4.21EE41 pKa = 5.12APTTGTKK48 pKa = 10.1HH49 pKa = 6.03IQGSWLKK56 pKa = 10.18AACQLIIEE64 pKa = 4.84GKK66 pKa = 7.86SEE68 pKa = 3.9RR69 pKa = 11.84SIAEE73 pKa = 3.92QFPSQYY79 pKa = 11.1VMYY82 pKa = 10.48HH83 pKa = 6.55RR84 pKa = 11.84GFQALRR90 pKa = 11.84NAIAPPKK97 pKa = 9.95PRR99 pKa = 11.84DD100 pKa = 3.58FKK102 pKa = 10.79TEE104 pKa = 3.78VYY106 pKa = 10.63VYY108 pKa = 10.17IGPTGTGKK116 pKa = 10.0SRR118 pKa = 11.84KK119 pKa = 7.78AAEE122 pKa = 4.17MATQTNEE129 pKa = 4.04PIFYY133 pKa = 10.08KK134 pKa = 10.8ARR136 pKa = 11.84GNWWDD141 pKa = 3.96GYY143 pKa = 9.77KK144 pKa = 9.85QQEE147 pKa = 4.13NVIIDD152 pKa = 4.13DD153 pKa = 4.46FYY155 pKa = 11.76GWLEE159 pKa = 3.89LDD161 pKa = 3.98EE162 pKa = 4.72LLKK165 pKa = 10.06ITDD168 pKa = 4.14RR169 pKa = 11.84YY170 pKa = 9.19PYY172 pKa = 9.57RR173 pKa = 11.84VPVKK177 pKa = 10.41GSYY180 pKa = 9.35EE181 pKa = 3.99VFNSKK186 pKa = 10.53RR187 pKa = 11.84IWITSNKK194 pKa = 8.46TVTTWYY200 pKa = 7.47PTEE203 pKa = 4.29RR204 pKa = 11.84DD205 pKa = 3.71EE206 pKa = 6.41LLLALTRR213 pKa = 11.84RR214 pKa = 11.84FTQIEE219 pKa = 4.56YY220 pKa = 10.58FF221 pKa = 3.66
MM1 pKa = 7.39EE2 pKa = 5.14RR3 pKa = 11.84VTLKK7 pKa = 10.63RR8 pKa = 11.84FCFTLNNYY16 pKa = 10.45DD17 pKa = 3.62EE18 pKa = 5.32DD19 pKa = 3.74RR20 pKa = 11.84VKK22 pKa = 10.42ILKK25 pKa = 10.11EE26 pKa = 3.99FMSTKK31 pKa = 9.84CKK33 pKa = 8.81YY34 pKa = 10.62AIMGFEE40 pKa = 4.21EE41 pKa = 5.12APTTGTKK48 pKa = 10.1HH49 pKa = 6.03IQGSWLKK56 pKa = 10.18AACQLIIEE64 pKa = 4.84GKK66 pKa = 7.86SEE68 pKa = 3.9RR69 pKa = 11.84SIAEE73 pKa = 3.92QFPSQYY79 pKa = 11.1VMYY82 pKa = 10.48HH83 pKa = 6.55RR84 pKa = 11.84GFQALRR90 pKa = 11.84NAIAPPKK97 pKa = 9.95PRR99 pKa = 11.84DD100 pKa = 3.58FKK102 pKa = 10.79TEE104 pKa = 3.78VYY106 pKa = 10.63VYY108 pKa = 10.17IGPTGTGKK116 pKa = 10.0SRR118 pKa = 11.84KK119 pKa = 7.78AAEE122 pKa = 4.17MATQTNEE129 pKa = 4.04PIFYY133 pKa = 10.08KK134 pKa = 10.8ARR136 pKa = 11.84GNWWDD141 pKa = 3.96GYY143 pKa = 9.77KK144 pKa = 9.85QQEE147 pKa = 4.13NVIIDD152 pKa = 4.13DD153 pKa = 4.46FYY155 pKa = 11.76GWLEE159 pKa = 3.89LDD161 pKa = 3.98EE162 pKa = 4.72LLKK165 pKa = 10.06ITDD168 pKa = 4.14RR169 pKa = 11.84YY170 pKa = 9.19PYY172 pKa = 9.57RR173 pKa = 11.84VPVKK177 pKa = 10.41GSYY180 pKa = 9.35EE181 pKa = 3.99VFNSKK186 pKa = 10.53RR187 pKa = 11.84IWITSNKK194 pKa = 8.46TVTTWYY200 pKa = 7.47PTEE203 pKa = 4.29RR204 pKa = 11.84DD205 pKa = 3.71EE206 pKa = 6.41LLLALTRR213 pKa = 11.84RR214 pKa = 11.84FTQIEE219 pKa = 4.56YY220 pKa = 10.58FF221 pKa = 3.66
Molecular weight: 26.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1N7TEH5|A0A1N7TEH5_9CIRC Replication-associated protein OS=Indian encephalitis associated cyclovirus OX=1755290 PE=4 SV=1
MM1 pKa = 6.49WRR3 pKa = 11.84ARR5 pKa = 11.84RR6 pKa = 11.84GRR8 pKa = 11.84YY9 pKa = 9.11RR10 pKa = 11.84YY11 pKa = 9.17VGRR14 pKa = 11.84AGVYY18 pKa = 8.87KK19 pKa = 10.3NRR21 pKa = 11.84RR22 pKa = 11.84IYY24 pKa = 10.07RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84YY29 pKa = 9.52RR30 pKa = 11.84LPLYY34 pKa = 10.07RR35 pKa = 11.84PQRR38 pKa = 11.84LRR40 pKa = 11.84RR41 pKa = 11.84STGLDD46 pKa = 3.03VYY48 pKa = 10.88AVKK51 pKa = 10.24TEE53 pKa = 4.31VIVPTLDD60 pKa = 3.65DD61 pKa = 3.68NVIVLEE67 pKa = 4.55PYY69 pKa = 10.01ISDD72 pKa = 3.85FRR74 pKa = 11.84RR75 pKa = 11.84LADD78 pKa = 3.48TYY80 pKa = 11.68NLYY83 pKa = 10.7NPYY86 pKa = 10.48RR87 pKa = 11.84FMSLSVRR94 pKa = 11.84VVPWCNTTSSTVPNNLYY111 pKa = 10.41CCAPFHH117 pKa = 7.09RR118 pKa = 11.84PLTMKK123 pKa = 10.35VNEE126 pKa = 4.02ATYY129 pKa = 10.69VQILSLPRR137 pKa = 11.84SKK139 pKa = 10.52SYY141 pKa = 9.97PATRR145 pKa = 11.84SSYY148 pKa = 10.27RR149 pKa = 11.84RR150 pKa = 11.84FVPTVASVLSEE161 pKa = 4.26TGQSTVLTGSGLTRR175 pKa = 11.84YY176 pKa = 9.63RR177 pKa = 11.84PLISQSPGYY186 pKa = 9.9DD187 pKa = 2.96AAAVRR192 pKa = 11.84HH193 pKa = 5.19YY194 pKa = 10.61CAIYY198 pKa = 10.22KK199 pKa = 8.66FSPPASGQPPVEE211 pKa = 4.03YY212 pKa = 10.01PLILRR217 pKa = 11.84AHH219 pKa = 5.81VRR221 pKa = 11.84FFSPKK226 pKa = 9.62DD227 pKa = 3.34TDD229 pKa = 3.96FSS231 pKa = 4.0
MM1 pKa = 6.49WRR3 pKa = 11.84ARR5 pKa = 11.84RR6 pKa = 11.84GRR8 pKa = 11.84YY9 pKa = 9.11RR10 pKa = 11.84YY11 pKa = 9.17VGRR14 pKa = 11.84AGVYY18 pKa = 8.87KK19 pKa = 10.3NRR21 pKa = 11.84RR22 pKa = 11.84IYY24 pKa = 10.07RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84YY29 pKa = 9.52RR30 pKa = 11.84LPLYY34 pKa = 10.07RR35 pKa = 11.84PQRR38 pKa = 11.84LRR40 pKa = 11.84RR41 pKa = 11.84STGLDD46 pKa = 3.03VYY48 pKa = 10.88AVKK51 pKa = 10.24TEE53 pKa = 4.31VIVPTLDD60 pKa = 3.65DD61 pKa = 3.68NVIVLEE67 pKa = 4.55PYY69 pKa = 10.01ISDD72 pKa = 3.85FRR74 pKa = 11.84RR75 pKa = 11.84LADD78 pKa = 3.48TYY80 pKa = 11.68NLYY83 pKa = 10.7NPYY86 pKa = 10.48RR87 pKa = 11.84FMSLSVRR94 pKa = 11.84VVPWCNTTSSTVPNNLYY111 pKa = 10.41CCAPFHH117 pKa = 7.09RR118 pKa = 11.84PLTMKK123 pKa = 10.35VNEE126 pKa = 4.02ATYY129 pKa = 10.69VQILSLPRR137 pKa = 11.84SKK139 pKa = 10.52SYY141 pKa = 9.97PATRR145 pKa = 11.84SSYY148 pKa = 10.27RR149 pKa = 11.84RR150 pKa = 11.84FVPTVASVLSEE161 pKa = 4.26TGQSTVLTGSGLTRR175 pKa = 11.84YY176 pKa = 9.63RR177 pKa = 11.84PLISQSPGYY186 pKa = 9.9DD187 pKa = 2.96AAAVRR192 pKa = 11.84HH193 pKa = 5.19YY194 pKa = 10.61CAIYY198 pKa = 10.22KK199 pKa = 8.66FSPPASGQPPVEE211 pKa = 4.03YY212 pKa = 10.01PLILRR217 pKa = 11.84AHH219 pKa = 5.81VRR221 pKa = 11.84FFSPKK226 pKa = 9.62DD227 pKa = 3.34TDD229 pKa = 3.96FSS231 pKa = 4.0
Molecular weight: 26.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
452 |
221 |
231 |
226.0 |
26.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.973 ± 0.058 | 1.549 ± 0.121 |
3.761 ± 0.197 | 5.088 ± 1.932 |
4.425 ± 0.635 | 4.646 ± 0.496 |
1.106 ± 0.127 | 5.088 ± 1.074 |
5.31 ± 1.792 | 7.301 ± 0.611 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.77 ± 0.311 | 3.54 ± 0.051 |
6.637 ± 1.335 | 3.097 ± 0.616 |
10.177 ± 2.143 | 6.637 ± 1.621 |
7.743 ± 0.54 | 6.858 ± 1.475 |
1.77 ± 0.597 | 7.522 ± 0.751 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
