
Potato yellow vein virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Closteroviridae; Crinivirus
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
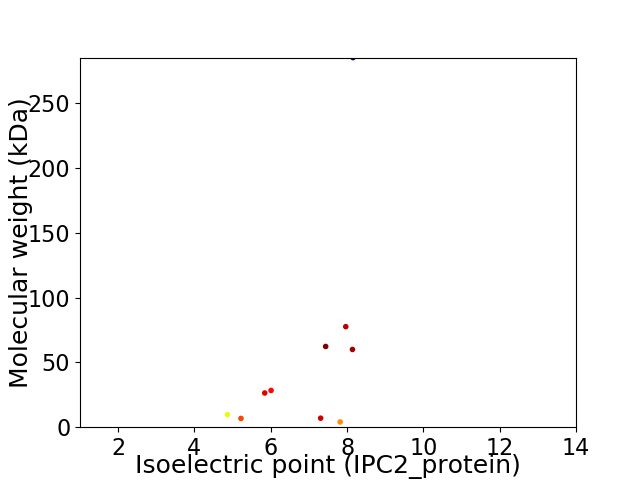
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q70PI3|Q70PI3_9CLOS Uncharacterized protein OS=Potato yellow vein virus OX=103881 PE=4 SV=1
MM1 pKa = 7.42NSISEE6 pKa = 4.89LITEE10 pKa = 4.71FGEE13 pKa = 4.43DD14 pKa = 3.08RR15 pKa = 11.84VDD17 pKa = 4.31EE18 pKa = 4.32IFALHH23 pKa = 6.91RR24 pKa = 11.84SFGQSNPPLCDD35 pKa = 3.03VLLNLISSKK44 pKa = 10.57LFSHH48 pKa = 7.48DD49 pKa = 3.8DD50 pKa = 3.5VFEE53 pKa = 3.94EE54 pKa = 4.9SIIEE58 pKa = 4.17FGDD61 pKa = 3.53LKK63 pKa = 11.3SFLLVTKK70 pKa = 8.56FIRR73 pKa = 11.84GVYY76 pKa = 8.84NSRR79 pKa = 11.84YY80 pKa = 7.51NLRR83 pKa = 11.84YY84 pKa = 9.72
MM1 pKa = 7.42NSISEE6 pKa = 4.89LITEE10 pKa = 4.71FGEE13 pKa = 4.43DD14 pKa = 3.08RR15 pKa = 11.84VDD17 pKa = 4.31EE18 pKa = 4.32IFALHH23 pKa = 6.91RR24 pKa = 11.84SFGQSNPPLCDD35 pKa = 3.03VLLNLISSKK44 pKa = 10.57LFSHH48 pKa = 7.48DD49 pKa = 3.8DD50 pKa = 3.5VFEE53 pKa = 3.94EE54 pKa = 4.9SIIEE58 pKa = 4.17FGDD61 pKa = 3.53LKK63 pKa = 11.3SFLLVTKK70 pKa = 8.56FIRR73 pKa = 11.84GVYY76 pKa = 8.84NSRR79 pKa = 11.84YY80 pKa = 7.51NLRR83 pKa = 11.84YY84 pKa = 9.72
Molecular weight: 9.75 kDa
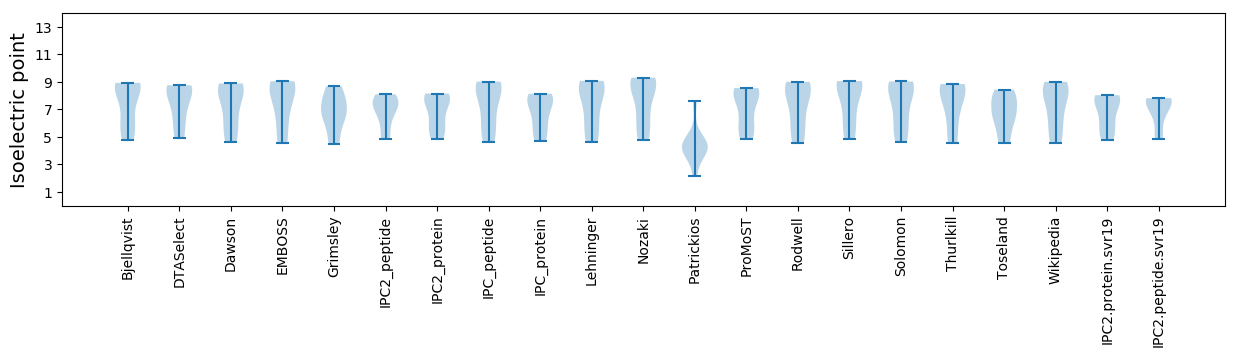
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q70PI4|Q70PI4_9CLOS p7 OS=Potato yellow vein virus OX=103881 PE=4 SV=1
MM1 pKa = 7.54VSIFRR6 pKa = 11.84SPVVRR11 pKa = 11.84DD12 pKa = 3.65FLSMVFKK19 pKa = 10.81TNDD22 pKa = 3.26PSEE25 pKa = 4.78KK26 pKa = 9.28IGEE29 pKa = 4.08INKK32 pKa = 9.48YY33 pKa = 8.84LVKK36 pKa = 10.63NFHH39 pKa = 7.02IEE41 pKa = 3.92NQNKK45 pKa = 9.06YY46 pKa = 7.97RR47 pKa = 11.84TNIKK51 pKa = 10.31GRR53 pKa = 11.84LTEE56 pKa = 3.89LTSNYY61 pKa = 9.01FVRR64 pKa = 11.84NNEE67 pKa = 3.53IFVNDD72 pKa = 3.79GNFLEE77 pKa = 4.32ILKK80 pKa = 10.61LPVIFMYY87 pKa = 10.66RR88 pKa = 11.84IRR90 pKa = 11.84PEE92 pKa = 3.81LTKK95 pKa = 10.96NSAYY99 pKa = 10.84LPVNIFASYY108 pKa = 10.83NYY110 pKa = 9.87EE111 pKa = 3.83FYY113 pKa = 11.01KK114 pKa = 10.99NSFSPHH120 pKa = 5.54YY121 pKa = 10.81DD122 pKa = 3.0KK123 pKa = 11.42DD124 pKa = 4.14LNSFLSSNKK133 pKa = 10.07EE134 pKa = 3.48IGCLYY139 pKa = 9.47TEE141 pKa = 4.73RR142 pKa = 11.84DD143 pKa = 3.52IQEE146 pKa = 4.86HH147 pKa = 5.9YY148 pKa = 10.51PSVHH152 pKa = 5.33GVRR155 pKa = 11.84LLTLYY160 pKa = 10.08RR161 pKa = 11.84VCNSLGRR168 pKa = 11.84YY169 pKa = 8.81VDD171 pKa = 5.25LNEE174 pKa = 5.15LDD176 pKa = 3.48NGNIKK181 pKa = 10.42GFSIDD186 pKa = 3.88PDD188 pKa = 3.8SKK190 pKa = 11.5AKK192 pKa = 10.39IIGEE196 pKa = 4.23GLSSNPLFNSCVEE209 pKa = 3.91IFRR212 pKa = 11.84SYY214 pKa = 11.63LEE216 pKa = 4.12LSATKK221 pKa = 10.42AGNAKK226 pKa = 9.93VEE228 pKa = 4.19ANKK231 pKa = 10.64KK232 pKa = 10.03IFTTYY237 pKa = 10.09LNCMSLGEE245 pKa = 4.21SYY247 pKa = 11.03EE248 pKa = 4.3RR249 pKa = 11.84IRR251 pKa = 11.84KK252 pKa = 8.68NPLVIAKK259 pKa = 8.84FIKK262 pKa = 10.38LYY264 pKa = 11.12EE265 pKa = 4.24EE266 pKa = 4.13FTKK269 pKa = 10.52EE270 pKa = 3.86SRR272 pKa = 11.84GFKK275 pKa = 10.51DD276 pKa = 3.2DD277 pKa = 4.09FEE279 pKa = 6.81AIKK282 pKa = 10.87LLTPRR287 pKa = 11.84FKK289 pKa = 11.24VFLKK293 pKa = 10.68KK294 pKa = 10.9VFDD297 pKa = 4.31VNSSLNDD304 pKa = 4.36DD305 pKa = 3.68ILFIEE310 pKa = 4.63MPKK313 pKa = 10.56SSVVDD318 pKa = 4.03LLSEE322 pKa = 3.98PVSVGEE328 pKa = 3.99YY329 pKa = 10.28LRR331 pKa = 11.84SKK333 pKa = 10.66DD334 pKa = 3.7AISVVSTSNTLSEE347 pKa = 4.43KK348 pKa = 10.86ADD350 pKa = 3.87LLATHH355 pKa = 7.4IILRR359 pKa = 11.84FLQDD363 pKa = 3.82KK364 pKa = 10.66LPAPEE369 pKa = 6.08DD370 pKa = 3.4ILIDD374 pKa = 3.96AFLMILGRR382 pKa = 11.84TTTNSKK388 pKa = 10.61RR389 pKa = 11.84FGKK392 pKa = 9.66PVLVTLKK399 pKa = 10.27YY400 pKa = 10.76EE401 pKa = 4.15NMSKK405 pKa = 10.51EE406 pKa = 3.47IDD408 pKa = 3.13ISVVVNALNNKK419 pKa = 9.07VSGAHH424 pKa = 6.02NEE426 pKa = 3.9YY427 pKa = 10.9LGMNVLRR434 pKa = 11.84LWANSRR440 pKa = 11.84SSRR443 pKa = 11.84AMRR446 pKa = 11.84LFRR449 pKa = 11.84SLNFNPGLFSYY460 pKa = 10.92CPGILDD466 pKa = 3.34YY467 pKa = 11.03MRR469 pKa = 11.84FDD471 pKa = 4.51FYY473 pKa = 11.53KK474 pKa = 10.77AIPLKK479 pKa = 11.08DD480 pKa = 3.41MTDD483 pKa = 3.48EE484 pKa = 4.22EE485 pKa = 4.69VQSFRR490 pKa = 11.84TLRR493 pKa = 11.84LHH495 pKa = 5.23TEE497 pKa = 3.73KK498 pKa = 10.34RR499 pKa = 11.84TDD501 pKa = 3.42PQGSYY506 pKa = 10.35SAEE509 pKa = 3.79CDD511 pKa = 3.01NWILNLKK518 pKa = 9.13NRR520 pKa = 11.84SS521 pKa = 3.4
MM1 pKa = 7.54VSIFRR6 pKa = 11.84SPVVRR11 pKa = 11.84DD12 pKa = 3.65FLSMVFKK19 pKa = 10.81TNDD22 pKa = 3.26PSEE25 pKa = 4.78KK26 pKa = 9.28IGEE29 pKa = 4.08INKK32 pKa = 9.48YY33 pKa = 8.84LVKK36 pKa = 10.63NFHH39 pKa = 7.02IEE41 pKa = 3.92NQNKK45 pKa = 9.06YY46 pKa = 7.97RR47 pKa = 11.84TNIKK51 pKa = 10.31GRR53 pKa = 11.84LTEE56 pKa = 3.89LTSNYY61 pKa = 9.01FVRR64 pKa = 11.84NNEE67 pKa = 3.53IFVNDD72 pKa = 3.79GNFLEE77 pKa = 4.32ILKK80 pKa = 10.61LPVIFMYY87 pKa = 10.66RR88 pKa = 11.84IRR90 pKa = 11.84PEE92 pKa = 3.81LTKK95 pKa = 10.96NSAYY99 pKa = 10.84LPVNIFASYY108 pKa = 10.83NYY110 pKa = 9.87EE111 pKa = 3.83FYY113 pKa = 11.01KK114 pKa = 10.99NSFSPHH120 pKa = 5.54YY121 pKa = 10.81DD122 pKa = 3.0KK123 pKa = 11.42DD124 pKa = 4.14LNSFLSSNKK133 pKa = 10.07EE134 pKa = 3.48IGCLYY139 pKa = 9.47TEE141 pKa = 4.73RR142 pKa = 11.84DD143 pKa = 3.52IQEE146 pKa = 4.86HH147 pKa = 5.9YY148 pKa = 10.51PSVHH152 pKa = 5.33GVRR155 pKa = 11.84LLTLYY160 pKa = 10.08RR161 pKa = 11.84VCNSLGRR168 pKa = 11.84YY169 pKa = 8.81VDD171 pKa = 5.25LNEE174 pKa = 5.15LDD176 pKa = 3.48NGNIKK181 pKa = 10.42GFSIDD186 pKa = 3.88PDD188 pKa = 3.8SKK190 pKa = 11.5AKK192 pKa = 10.39IIGEE196 pKa = 4.23GLSSNPLFNSCVEE209 pKa = 3.91IFRR212 pKa = 11.84SYY214 pKa = 11.63LEE216 pKa = 4.12LSATKK221 pKa = 10.42AGNAKK226 pKa = 9.93VEE228 pKa = 4.19ANKK231 pKa = 10.64KK232 pKa = 10.03IFTTYY237 pKa = 10.09LNCMSLGEE245 pKa = 4.21SYY247 pKa = 11.03EE248 pKa = 4.3RR249 pKa = 11.84IRR251 pKa = 11.84KK252 pKa = 8.68NPLVIAKK259 pKa = 8.84FIKK262 pKa = 10.38LYY264 pKa = 11.12EE265 pKa = 4.24EE266 pKa = 4.13FTKK269 pKa = 10.52EE270 pKa = 3.86SRR272 pKa = 11.84GFKK275 pKa = 10.51DD276 pKa = 3.2DD277 pKa = 4.09FEE279 pKa = 6.81AIKK282 pKa = 10.87LLTPRR287 pKa = 11.84FKK289 pKa = 11.24VFLKK293 pKa = 10.68KK294 pKa = 10.9VFDD297 pKa = 4.31VNSSLNDD304 pKa = 4.36DD305 pKa = 3.68ILFIEE310 pKa = 4.63MPKK313 pKa = 10.56SSVVDD318 pKa = 4.03LLSEE322 pKa = 3.98PVSVGEE328 pKa = 3.99YY329 pKa = 10.28LRR331 pKa = 11.84SKK333 pKa = 10.66DD334 pKa = 3.7AISVVSTSNTLSEE347 pKa = 4.43KK348 pKa = 10.86ADD350 pKa = 3.87LLATHH355 pKa = 7.4IILRR359 pKa = 11.84FLQDD363 pKa = 3.82KK364 pKa = 10.66LPAPEE369 pKa = 6.08DD370 pKa = 3.4ILIDD374 pKa = 3.96AFLMILGRR382 pKa = 11.84TTTNSKK388 pKa = 10.61RR389 pKa = 11.84FGKK392 pKa = 9.66PVLVTLKK399 pKa = 10.27YY400 pKa = 10.76EE401 pKa = 4.15NMSKK405 pKa = 10.51EE406 pKa = 3.47IDD408 pKa = 3.13ISVVVNALNNKK419 pKa = 9.07VSGAHH424 pKa = 6.02NEE426 pKa = 3.9YY427 pKa = 10.9LGMNVLRR434 pKa = 11.84LWANSRR440 pKa = 11.84SSRR443 pKa = 11.84AMRR446 pKa = 11.84LFRR449 pKa = 11.84SLNFNPGLFSYY460 pKa = 10.92CPGILDD466 pKa = 3.34YY467 pKa = 11.03MRR469 pKa = 11.84FDD471 pKa = 4.51FYY473 pKa = 11.53KK474 pKa = 10.77AIPLKK479 pKa = 11.08DD480 pKa = 3.41MTDD483 pKa = 3.48EE484 pKa = 4.22EE485 pKa = 4.69VQSFRR490 pKa = 11.84TLRR493 pKa = 11.84LHH495 pKa = 5.23TEE497 pKa = 3.73KK498 pKa = 10.34RR499 pKa = 11.84TDD501 pKa = 3.42PQGSYY506 pKa = 10.35SAEE509 pKa = 3.79CDD511 pKa = 3.01NWILNLKK518 pKa = 9.13NRR520 pKa = 11.84SS521 pKa = 3.4
Molecular weight: 60.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4944 |
35 |
2475 |
494.4 |
56.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.236 ± 0.32 | 1.82 ± 0.197 |
6.614 ± 0.333 | 5.643 ± 0.311 |
5.542 ± 0.348 | 4.632 ± 0.228 |
1.517 ± 0.089 | 7.201 ± 0.254 |
8.212 ± 0.287 | 9.79 ± 0.461 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.488 ± 0.311 | 6.472 ± 0.508 |
3.196 ± 0.249 | 2.447 ± 0.457 |
5.34 ± 0.386 | 9.244 ± 0.25 |
4.693 ± 0.37 | 7.019 ± 0.225 |
0.445 ± 0.089 | 4.45 ± 0.308 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
