
Athtab bunya-like virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; unclassified Bunyavirales
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
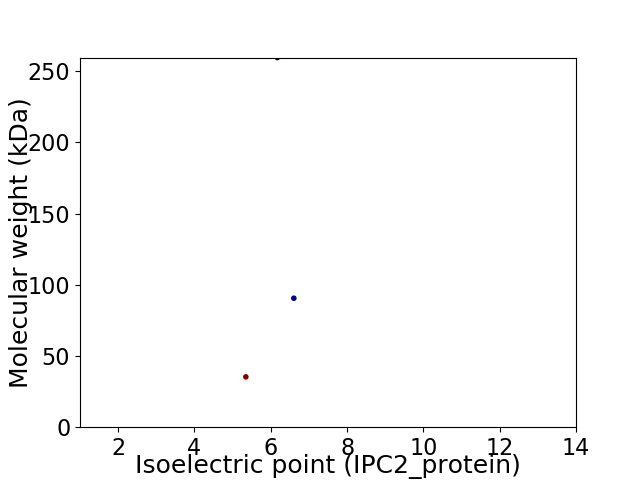
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3G1WQW8|A0A3G1WQW8_9VIRU Glycoprotein C OS=Athtab bunya-like virus OX=2315724 PE=4 SV=1
MM1 pKa = 7.58LKK3 pKa = 9.25TVFLLLTSTTLLVDD17 pKa = 3.41NAVVMTSMGGRR28 pKa = 11.84YY29 pKa = 9.01RR30 pKa = 11.84LCFGSFCYY38 pKa = 10.58YY39 pKa = 10.71SEE41 pKa = 3.83FAAYY45 pKa = 9.44KK46 pKa = 9.97EE47 pKa = 4.05QFEE50 pKa = 4.23AQAAFQCAHH59 pKa = 7.1LLANIVAIDD68 pKa = 4.01TTTSTCSVYY77 pKa = 10.58KK78 pKa = 10.46PRR80 pKa = 11.84ACFAQVEE87 pKa = 4.52QGEE90 pKa = 4.74GPYY93 pKa = 10.29LVPFDD98 pKa = 3.91YY99 pKa = 10.93TILQSTNFIPAHH111 pKa = 6.39LGCITYY117 pKa = 9.42TSSQNTFAYY126 pKa = 6.92KK127 pKa = 9.6TCSTPSALKK136 pKa = 10.45DD137 pKa = 3.47PSVVVPITTTTTTFSFTVYY156 pKa = 10.71KK157 pKa = 10.52DD158 pKa = 3.74YY159 pKa = 10.88TCEE162 pKa = 3.94AVLSPTWTTTVWTDD176 pKa = 2.9TKK178 pKa = 10.84IVITSTIVTNLITSITVVNCTITNTHH204 pKa = 6.86LSIQGYY210 pKa = 6.75TLTTTEE216 pKa = 4.69LLLTTALAKK225 pKa = 10.19PAVTNITSVTFTTTQLNIVNHH246 pKa = 5.93VLTTTSVLFTEE257 pKa = 4.56TTYY260 pKa = 10.89TITEE264 pKa = 3.94YY265 pKa = 10.81HH266 pKa = 6.14HH267 pKa = 6.61TVTHH271 pKa = 5.94ADD273 pKa = 3.5EE274 pKa = 4.77YY275 pKa = 10.44KK276 pKa = 10.15TLEE279 pKa = 4.15PVLDD283 pKa = 3.82RR284 pKa = 11.84KK285 pKa = 8.23TCNLLGMHH293 pKa = 6.87FCDD296 pKa = 5.24KK297 pKa = 10.57CLPPNKK303 pKa = 9.13ICNGIKK309 pKa = 10.32DD310 pKa = 4.22CADD313 pKa = 3.47GTDD316 pKa = 3.83EE317 pKa = 5.48PSNCC321 pKa = 4.38
MM1 pKa = 7.58LKK3 pKa = 9.25TVFLLLTSTTLLVDD17 pKa = 3.41NAVVMTSMGGRR28 pKa = 11.84YY29 pKa = 9.01RR30 pKa = 11.84LCFGSFCYY38 pKa = 10.58YY39 pKa = 10.71SEE41 pKa = 3.83FAAYY45 pKa = 9.44KK46 pKa = 9.97EE47 pKa = 4.05QFEE50 pKa = 4.23AQAAFQCAHH59 pKa = 7.1LLANIVAIDD68 pKa = 4.01TTTSTCSVYY77 pKa = 10.58KK78 pKa = 10.46PRR80 pKa = 11.84ACFAQVEE87 pKa = 4.52QGEE90 pKa = 4.74GPYY93 pKa = 10.29LVPFDD98 pKa = 3.91YY99 pKa = 10.93TILQSTNFIPAHH111 pKa = 6.39LGCITYY117 pKa = 9.42TSSQNTFAYY126 pKa = 6.92KK127 pKa = 9.6TCSTPSALKK136 pKa = 10.45DD137 pKa = 3.47PSVVVPITTTTTTFSFTVYY156 pKa = 10.71KK157 pKa = 10.52DD158 pKa = 3.74YY159 pKa = 10.88TCEE162 pKa = 3.94AVLSPTWTTTVWTDD176 pKa = 2.9TKK178 pKa = 10.84IVITSTIVTNLITSITVVNCTITNTHH204 pKa = 6.86LSIQGYY210 pKa = 6.75TLTTTEE216 pKa = 4.69LLLTTALAKK225 pKa = 10.19PAVTNITSVTFTTTQLNIVNHH246 pKa = 5.93VLTTTSVLFTEE257 pKa = 4.56TTYY260 pKa = 10.89TITEE264 pKa = 3.94YY265 pKa = 10.81HH266 pKa = 6.14HH267 pKa = 6.61TVTHH271 pKa = 5.94ADD273 pKa = 3.5EE274 pKa = 4.77YY275 pKa = 10.44KK276 pKa = 10.15TLEE279 pKa = 4.15PVLDD283 pKa = 3.82RR284 pKa = 11.84KK285 pKa = 8.23TCNLLGMHH293 pKa = 6.87FCDD296 pKa = 5.24KK297 pKa = 10.57CLPPNKK303 pKa = 9.13ICNGIKK309 pKa = 10.32DD310 pKa = 4.22CADD313 pKa = 3.47GTDD316 pKa = 3.83EE317 pKa = 5.48PSNCC321 pKa = 4.38
Molecular weight: 35.35 kDa
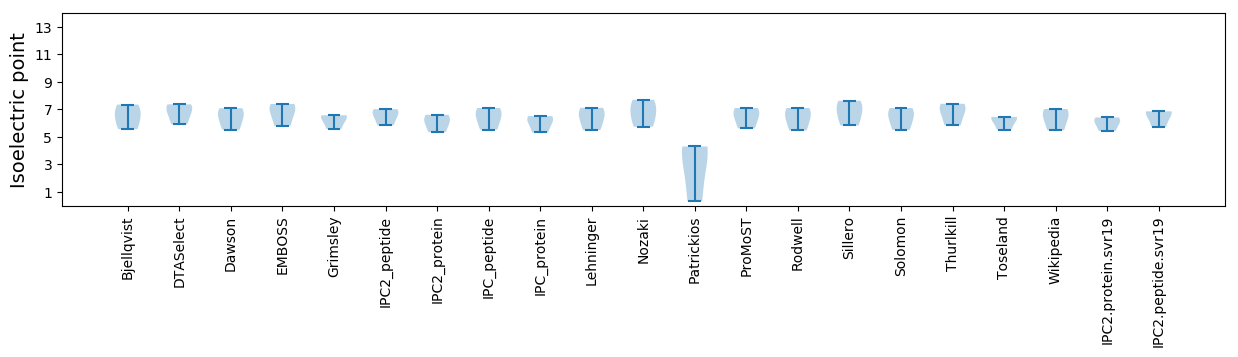
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G1WQW8|A0A3G1WQW8_9VIRU Glycoprotein C OS=Athtab bunya-like virus OX=2315724 PE=4 SV=1
MM1 pKa = 7.72RR2 pKa = 11.84NILEE6 pKa = 4.24KK7 pKa = 10.87SSMYY11 pKa = 10.17MLILLFVEE19 pKa = 5.02LVVSSGVFVKK29 pKa = 10.05TNNCTSLEE37 pKa = 4.03KK38 pKa = 10.59PQDD41 pKa = 3.6CEE43 pKa = 3.99EE44 pKa = 4.11FGEE47 pKa = 4.35LGGVHH52 pKa = 6.06WARR55 pKa = 11.84LKK57 pKa = 10.86DD58 pKa = 3.64RR59 pKa = 11.84QDD61 pKa = 3.06KK62 pKa = 11.45YY63 pKa = 11.13EE64 pKa = 4.05IQAKK68 pKa = 10.38LSGPVCTGFSDD79 pKa = 5.29PILIGDD85 pKa = 3.7SRR87 pKa = 11.84FSYY90 pKa = 10.82SVDD93 pKa = 3.03GQFRR97 pKa = 11.84SLSGEE102 pKa = 4.04VVDD105 pKa = 5.43RR106 pKa = 11.84CSQVEE111 pKa = 3.83NDD113 pKa = 4.52HH114 pKa = 6.9FPEE117 pKa = 4.98CGTAVVSVCHH127 pKa = 7.08LIPIGNCEE135 pKa = 4.2LKK137 pKa = 10.73VDD139 pKa = 4.39LEE141 pKa = 4.32SSKK144 pKa = 10.26VHH146 pKa = 5.84VSGKK150 pKa = 8.94TSGPLQYY157 pKa = 10.51VIQINEE163 pKa = 3.99EE164 pKa = 3.59VDD166 pKa = 3.73MEE168 pKa = 4.35GHH170 pKa = 6.69CLNCEE175 pKa = 4.1GFSRR179 pKa = 11.84SYY181 pKa = 10.55PLQEE185 pKa = 4.42GLTTYY190 pKa = 10.94HH191 pKa = 6.28LTCNRR196 pKa = 11.84NSLSKK201 pKa = 9.58TLSLSKK207 pKa = 10.91SEE209 pKa = 4.0VCARR213 pKa = 11.84NAQDD217 pKa = 4.13FYY219 pKa = 11.73LGFHH223 pKa = 6.41YY224 pKa = 9.54KK225 pKa = 8.93WCMNAFWYY233 pKa = 7.7KK234 pKa = 9.74TLTILFFLVMLTACVPMINWPIHH257 pKa = 5.46MILNKK262 pKa = 10.4LIRR265 pKa = 11.84CCSRR269 pKa = 11.84SEE271 pKa = 4.08AKK273 pKa = 10.74VKK275 pKa = 8.59VTKK278 pKa = 10.36AVPSFLFLIWWVVLLSHH295 pKa = 6.93IVYY298 pKa = 10.71AEE300 pKa = 3.92ASLADD305 pKa = 3.63SLKK308 pKa = 9.92EE309 pKa = 3.77QGFKK313 pKa = 10.97PEE315 pKa = 4.01TVDD318 pKa = 4.49FKK320 pKa = 11.37GLYY323 pKa = 7.78RR324 pKa = 11.84TTVPEE329 pKa = 4.24ADD331 pKa = 3.99FEE333 pKa = 5.22LISTGTGEE341 pKa = 4.23ALRR344 pKa = 11.84VEE346 pKa = 4.36SFDD349 pKa = 3.64IKK351 pKa = 10.84LSPGYY356 pKa = 10.79ALGLVVDD363 pKa = 4.4NVEE366 pKa = 4.56LDD368 pKa = 3.96LKK370 pKa = 10.18VKK372 pKa = 9.63SVRR375 pKa = 11.84AQMSYY380 pKa = 10.74KK381 pKa = 10.34DD382 pKa = 4.23CYY384 pKa = 9.81STGDD388 pKa = 3.26TKK390 pKa = 10.69IISKK394 pKa = 8.5STDD397 pKa = 2.9TCTEE401 pKa = 3.84SCDD404 pKa = 3.44KK405 pKa = 10.85CVEE408 pKa = 4.09EE409 pKa = 4.87LVVPEE414 pKa = 4.05QVLPEE419 pKa = 4.12NVVKK423 pKa = 10.56VAKK426 pKa = 10.02SSSWACDD433 pKa = 3.21GAGCLSMATGCTCGYY448 pKa = 10.73CYY450 pKa = 7.56MTLTGPVWKK459 pKa = 10.78VCDD462 pKa = 3.88LEE464 pKa = 4.3LEE466 pKa = 4.15KK467 pKa = 11.32VEE469 pKa = 5.64INLCFNIGGKK479 pKa = 9.32GMCKK483 pKa = 9.06WLEE486 pKa = 4.15SVEE489 pKa = 4.23TFADD493 pKa = 3.58GLYY496 pKa = 10.36QINLKK501 pKa = 5.61TTKK504 pKa = 9.56IVLPEE509 pKa = 3.85RR510 pKa = 11.84VAEE513 pKa = 4.12VMEE516 pKa = 4.38EE517 pKa = 3.98GKK519 pKa = 10.51KK520 pKa = 9.42IRR522 pKa = 11.84KK523 pKa = 8.2IGLMNKK529 pKa = 10.02LGEE532 pKa = 4.22FDD534 pKa = 4.82SEE536 pKa = 4.41FGDD539 pKa = 3.9FQVVDD544 pKa = 3.61AGSNYY549 pKa = 10.12EE550 pKa = 3.99KK551 pKa = 10.9DD552 pKa = 3.05LDD554 pKa = 4.45YY555 pKa = 11.23IHH557 pKa = 7.07EE558 pKa = 4.37CHH560 pKa = 5.76FTQHH564 pKa = 5.8RR565 pKa = 11.84RR566 pKa = 11.84ITFKK570 pKa = 10.99SCGQNHH576 pKa = 6.5FYY578 pKa = 10.41KK579 pKa = 10.56HH580 pKa = 6.36KK581 pKa = 10.37NLHH584 pKa = 5.9PCQTCFDD591 pKa = 4.22GAKK594 pKa = 10.75GMDD597 pKa = 3.85FVDD600 pKa = 4.44RR601 pKa = 11.84PIGEE605 pKa = 4.37AQLKK609 pKa = 10.61LNLPLLGLSLKK620 pKa = 9.76EE621 pKa = 4.29DD622 pKa = 3.43KK623 pKa = 11.32VDD625 pKa = 3.4VSKK628 pKa = 11.12FKK630 pKa = 10.7ISSCKK635 pKa = 9.99GCRR638 pKa = 11.84NCQEE642 pKa = 3.87GMACKK647 pKa = 10.21ISFEE651 pKa = 4.44TKK653 pKa = 9.3QSGRR657 pKa = 11.84LPIKK661 pKa = 10.3CDD663 pKa = 3.12SAQSRR668 pKa = 11.84NYY670 pKa = 10.62LLAKK674 pKa = 9.79IGKK677 pKa = 6.57NTLMMTFFTDD687 pKa = 3.69LKK689 pKa = 10.81EE690 pKa = 4.34GVLKK694 pKa = 11.14CEE696 pKa = 4.21LGMLKK701 pKa = 10.07SQVNFKK707 pKa = 9.75TEE709 pKa = 3.99PGKK712 pKa = 10.86LLLEE716 pKa = 4.42DD717 pKa = 3.93FRR719 pKa = 11.84RR720 pKa = 11.84AVDD723 pKa = 3.47RR724 pKa = 11.84SLEE727 pKa = 4.02EE728 pKa = 5.71DD729 pKa = 3.78YY730 pKa = 11.13HH731 pKa = 7.11CQLFSCTMGRR741 pKa = 11.84FLYY744 pKa = 10.77NVFGLKK750 pKa = 10.42GLWHH754 pKa = 5.98YY755 pKa = 10.97LWIIFIFVILLVVVVKK771 pKa = 10.19LVKK774 pKa = 10.63LILNRR779 pKa = 11.84ATKK782 pKa = 10.49RR783 pKa = 11.84SMPIFKK789 pKa = 10.12DD790 pKa = 3.16SRR792 pKa = 11.84FFKK795 pKa = 9.72PQSVNVRR802 pKa = 3.49
MM1 pKa = 7.72RR2 pKa = 11.84NILEE6 pKa = 4.24KK7 pKa = 10.87SSMYY11 pKa = 10.17MLILLFVEE19 pKa = 5.02LVVSSGVFVKK29 pKa = 10.05TNNCTSLEE37 pKa = 4.03KK38 pKa = 10.59PQDD41 pKa = 3.6CEE43 pKa = 3.99EE44 pKa = 4.11FGEE47 pKa = 4.35LGGVHH52 pKa = 6.06WARR55 pKa = 11.84LKK57 pKa = 10.86DD58 pKa = 3.64RR59 pKa = 11.84QDD61 pKa = 3.06KK62 pKa = 11.45YY63 pKa = 11.13EE64 pKa = 4.05IQAKK68 pKa = 10.38LSGPVCTGFSDD79 pKa = 5.29PILIGDD85 pKa = 3.7SRR87 pKa = 11.84FSYY90 pKa = 10.82SVDD93 pKa = 3.03GQFRR97 pKa = 11.84SLSGEE102 pKa = 4.04VVDD105 pKa = 5.43RR106 pKa = 11.84CSQVEE111 pKa = 3.83NDD113 pKa = 4.52HH114 pKa = 6.9FPEE117 pKa = 4.98CGTAVVSVCHH127 pKa = 7.08LIPIGNCEE135 pKa = 4.2LKK137 pKa = 10.73VDD139 pKa = 4.39LEE141 pKa = 4.32SSKK144 pKa = 10.26VHH146 pKa = 5.84VSGKK150 pKa = 8.94TSGPLQYY157 pKa = 10.51VIQINEE163 pKa = 3.99EE164 pKa = 3.59VDD166 pKa = 3.73MEE168 pKa = 4.35GHH170 pKa = 6.69CLNCEE175 pKa = 4.1GFSRR179 pKa = 11.84SYY181 pKa = 10.55PLQEE185 pKa = 4.42GLTTYY190 pKa = 10.94HH191 pKa = 6.28LTCNRR196 pKa = 11.84NSLSKK201 pKa = 9.58TLSLSKK207 pKa = 10.91SEE209 pKa = 4.0VCARR213 pKa = 11.84NAQDD217 pKa = 4.13FYY219 pKa = 11.73LGFHH223 pKa = 6.41YY224 pKa = 9.54KK225 pKa = 8.93WCMNAFWYY233 pKa = 7.7KK234 pKa = 9.74TLTILFFLVMLTACVPMINWPIHH257 pKa = 5.46MILNKK262 pKa = 10.4LIRR265 pKa = 11.84CCSRR269 pKa = 11.84SEE271 pKa = 4.08AKK273 pKa = 10.74VKK275 pKa = 8.59VTKK278 pKa = 10.36AVPSFLFLIWWVVLLSHH295 pKa = 6.93IVYY298 pKa = 10.71AEE300 pKa = 3.92ASLADD305 pKa = 3.63SLKK308 pKa = 9.92EE309 pKa = 3.77QGFKK313 pKa = 10.97PEE315 pKa = 4.01TVDD318 pKa = 4.49FKK320 pKa = 11.37GLYY323 pKa = 7.78RR324 pKa = 11.84TTVPEE329 pKa = 4.24ADD331 pKa = 3.99FEE333 pKa = 5.22LISTGTGEE341 pKa = 4.23ALRR344 pKa = 11.84VEE346 pKa = 4.36SFDD349 pKa = 3.64IKK351 pKa = 10.84LSPGYY356 pKa = 10.79ALGLVVDD363 pKa = 4.4NVEE366 pKa = 4.56LDD368 pKa = 3.96LKK370 pKa = 10.18VKK372 pKa = 9.63SVRR375 pKa = 11.84AQMSYY380 pKa = 10.74KK381 pKa = 10.34DD382 pKa = 4.23CYY384 pKa = 9.81STGDD388 pKa = 3.26TKK390 pKa = 10.69IISKK394 pKa = 8.5STDD397 pKa = 2.9TCTEE401 pKa = 3.84SCDD404 pKa = 3.44KK405 pKa = 10.85CVEE408 pKa = 4.09EE409 pKa = 4.87LVVPEE414 pKa = 4.05QVLPEE419 pKa = 4.12NVVKK423 pKa = 10.56VAKK426 pKa = 10.02SSSWACDD433 pKa = 3.21GAGCLSMATGCTCGYY448 pKa = 10.73CYY450 pKa = 7.56MTLTGPVWKK459 pKa = 10.78VCDD462 pKa = 3.88LEE464 pKa = 4.3LEE466 pKa = 4.15KK467 pKa = 11.32VEE469 pKa = 5.64INLCFNIGGKK479 pKa = 9.32GMCKK483 pKa = 9.06WLEE486 pKa = 4.15SVEE489 pKa = 4.23TFADD493 pKa = 3.58GLYY496 pKa = 10.36QINLKK501 pKa = 5.61TTKK504 pKa = 9.56IVLPEE509 pKa = 3.85RR510 pKa = 11.84VAEE513 pKa = 4.12VMEE516 pKa = 4.38EE517 pKa = 3.98GKK519 pKa = 10.51KK520 pKa = 9.42IRR522 pKa = 11.84KK523 pKa = 8.2IGLMNKK529 pKa = 10.02LGEE532 pKa = 4.22FDD534 pKa = 4.82SEE536 pKa = 4.41FGDD539 pKa = 3.9FQVVDD544 pKa = 3.61AGSNYY549 pKa = 10.12EE550 pKa = 3.99KK551 pKa = 10.9DD552 pKa = 3.05LDD554 pKa = 4.45YY555 pKa = 11.23IHH557 pKa = 7.07EE558 pKa = 4.37CHH560 pKa = 5.76FTQHH564 pKa = 5.8RR565 pKa = 11.84RR566 pKa = 11.84ITFKK570 pKa = 10.99SCGQNHH576 pKa = 6.5FYY578 pKa = 10.41KK579 pKa = 10.56HH580 pKa = 6.36KK581 pKa = 10.37NLHH584 pKa = 5.9PCQTCFDD591 pKa = 4.22GAKK594 pKa = 10.75GMDD597 pKa = 3.85FVDD600 pKa = 4.44RR601 pKa = 11.84PIGEE605 pKa = 4.37AQLKK609 pKa = 10.61LNLPLLGLSLKK620 pKa = 9.76EE621 pKa = 4.29DD622 pKa = 3.43KK623 pKa = 11.32VDD625 pKa = 3.4VSKK628 pKa = 11.12FKK630 pKa = 10.7ISSCKK635 pKa = 9.99GCRR638 pKa = 11.84NCQEE642 pKa = 3.87GMACKK647 pKa = 10.21ISFEE651 pKa = 4.44TKK653 pKa = 9.3QSGRR657 pKa = 11.84LPIKK661 pKa = 10.3CDD663 pKa = 3.12SAQSRR668 pKa = 11.84NYY670 pKa = 10.62LLAKK674 pKa = 9.79IGKK677 pKa = 6.57NTLMMTFFTDD687 pKa = 3.69LKK689 pKa = 10.81EE690 pKa = 4.34GVLKK694 pKa = 11.14CEE696 pKa = 4.21LGMLKK701 pKa = 10.07SQVNFKK707 pKa = 9.75TEE709 pKa = 3.99PGKK712 pKa = 10.86LLLEE716 pKa = 4.42DD717 pKa = 3.93FRR719 pKa = 11.84RR720 pKa = 11.84AVDD723 pKa = 3.47RR724 pKa = 11.84SLEE727 pKa = 4.02EE728 pKa = 5.71DD729 pKa = 3.78YY730 pKa = 11.13HH731 pKa = 7.11CQLFSCTMGRR741 pKa = 11.84FLYY744 pKa = 10.77NVFGLKK750 pKa = 10.42GLWHH754 pKa = 5.98YY755 pKa = 10.97LWIIFIFVILLVVVVKK771 pKa = 10.19LVKK774 pKa = 10.63LILNRR779 pKa = 11.84ATKK782 pKa = 10.49RR783 pKa = 11.84SMPIFKK789 pKa = 10.12DD790 pKa = 3.16SRR792 pKa = 11.84FFKK795 pKa = 9.72PQSVNVRR802 pKa = 3.49
Molecular weight: 90.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3358 |
321 |
2235 |
1119.3 |
128.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.318 ± 0.322 | 2.74 ± 1.053 |
5.956 ± 0.669 | 6.82 ± 0.55 |
5.092 ± 0.102 | 4.795 ± 0.658 |
2.144 ± 0.059 | 6.015 ± 0.5 |
7.683 ± 0.595 | 10.125 ± 0.271 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.008 ± 0.411 | 5.152 ± 0.643 |
3.008 ± 0.186 | 3.127 ± 0.095 |
4.586 ± 0.811 | 7.385 ± 0.194 |
5.926 ± 2.335 | 6.968 ± 0.636 |
1.37 ± 0.135 | 3.782 ± 0.287 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
