
Bovine polyomavirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; unclassified Polyomaviridae
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
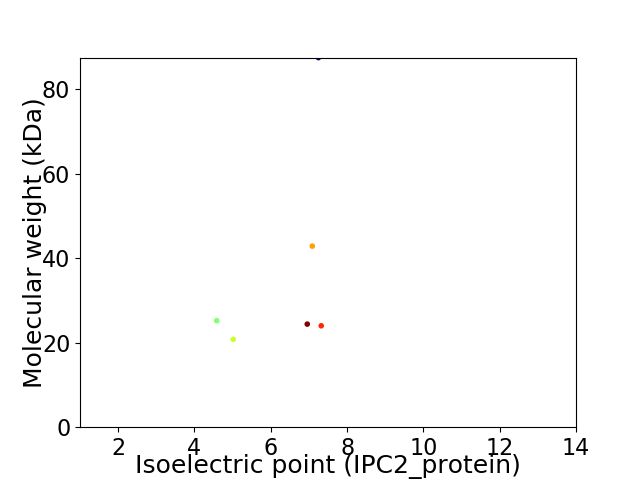
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A089QEX6|A0A089QEX6_9POLY Small T antigen OS=Bovine polyomavirus 2 OX=1578134 PE=4 SV=1
MM1 pKa = 7.41GAFISLLLNVGALAAEE17 pKa = 4.99LSAEE21 pKa = 4.17TAISVEE27 pKa = 4.41SILTGEE33 pKa = 4.25ALAALEE39 pKa = 4.35AEE41 pKa = 4.71IASVMTIQGISGIEE55 pKa = 4.01ALTQLGFTAEE65 pKa = 4.07QFSNFSLVSSLVNQALQYY83 pKa = 11.25AFYY86 pKa = 9.41FQTVSGAAALVEE98 pKa = 5.16AGIKK102 pKa = 8.89WGLGEE107 pKa = 4.34RR108 pKa = 11.84SVVDD112 pKa = 3.86ASDD115 pKa = 3.58VGLSPNGRR123 pKa = 11.84LLRR126 pKa = 11.84EE127 pKa = 3.7FAKK130 pKa = 10.59GFKK133 pKa = 9.41IDD135 pKa = 3.85PFNWSQSLIHH145 pKa = 6.56NIGRR149 pKa = 11.84TIFEE153 pKa = 5.28KK154 pKa = 10.53ISAHH158 pKa = 5.83NKK160 pKa = 6.66ITQATAQNYY169 pKa = 9.05ARR171 pKa = 11.84LIEE174 pKa = 3.88MGRR177 pKa = 11.84WYY179 pKa = 10.87VEE181 pKa = 4.04STDD184 pKa = 4.33SVTAGNDD191 pKa = 3.06SGLSVAVYY199 pKa = 7.28PAPGGAEE206 pKa = 3.79QRR208 pKa = 11.84ATPDD212 pKa = 2.71WLLPLILGLSGDD224 pKa = 4.0TTPTWKK230 pKa = 10.5ADD232 pKa = 3.58LEE234 pKa = 4.77INGSS238 pKa = 3.49
MM1 pKa = 7.41GAFISLLLNVGALAAEE17 pKa = 4.99LSAEE21 pKa = 4.17TAISVEE27 pKa = 4.41SILTGEE33 pKa = 4.25ALAALEE39 pKa = 4.35AEE41 pKa = 4.71IASVMTIQGISGIEE55 pKa = 4.01ALTQLGFTAEE65 pKa = 4.07QFSNFSLVSSLVNQALQYY83 pKa = 11.25AFYY86 pKa = 9.41FQTVSGAAALVEE98 pKa = 5.16AGIKK102 pKa = 8.89WGLGEE107 pKa = 4.34RR108 pKa = 11.84SVVDD112 pKa = 3.86ASDD115 pKa = 3.58VGLSPNGRR123 pKa = 11.84LLRR126 pKa = 11.84EE127 pKa = 3.7FAKK130 pKa = 10.59GFKK133 pKa = 9.41IDD135 pKa = 3.85PFNWSQSLIHH145 pKa = 6.56NIGRR149 pKa = 11.84TIFEE153 pKa = 5.28KK154 pKa = 10.53ISAHH158 pKa = 5.83NKK160 pKa = 6.66ITQATAQNYY169 pKa = 9.05ARR171 pKa = 11.84LIEE174 pKa = 3.88MGRR177 pKa = 11.84WYY179 pKa = 10.87VEE181 pKa = 4.04STDD184 pKa = 4.33SVTAGNDD191 pKa = 3.06SGLSVAVYY199 pKa = 7.28PAPGGAEE206 pKa = 3.79QRR208 pKa = 11.84ATPDD212 pKa = 2.71WLLPLILGLSGDD224 pKa = 4.0TTPTWKK230 pKa = 10.5ADD232 pKa = 3.58LEE234 pKa = 4.77INGSS238 pKa = 3.49
Molecular weight: 25.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A089RS32|A0A089RS32_9POLY VP1 OS=Bovine polyomavirus 2 OX=1578134 PE=3 SV=1
MM1 pKa = 7.99DD2 pKa = 6.15SILSRR7 pKa = 11.84QEE9 pKa = 3.63KK10 pKa = 10.49RR11 pKa = 11.84EE12 pKa = 3.98LCDD15 pKa = 3.54LLEE18 pKa = 4.82IPHH21 pKa = 6.66HH22 pKa = 6.83CYY24 pKa = 11.18GNIPMMKK31 pKa = 9.43AQYY34 pKa = 11.28KK35 pKa = 9.96KK36 pKa = 9.57MCLLYY41 pKa = 10.77HH42 pKa = 6.92PDD44 pKa = 3.64KK45 pKa = 11.46GGDD48 pKa = 3.38GAKK51 pKa = 9.02MVKK54 pKa = 10.2LNSLWTSFQDD64 pKa = 3.28EE65 pKa = 4.63VTKK68 pKa = 10.92LRR70 pKa = 11.84AEE72 pKa = 4.3VNFFSYY78 pKa = 10.14QVSAKK83 pKa = 9.8FFWDD87 pKa = 3.45LDD89 pKa = 3.87FNTLKK94 pKa = 10.68EE95 pKa = 4.18YY96 pKa = 10.72LGKK99 pKa = 9.76NQCKK103 pKa = 9.56FLRR106 pKa = 11.84GPNCLNSKK114 pKa = 10.22YY115 pKa = 10.34SYY117 pKa = 10.1CRR119 pKa = 11.84CIVCRR124 pKa = 11.84LSQQHH129 pKa = 5.93QDD131 pKa = 3.07IKK133 pKa = 10.81DD134 pKa = 3.75TQEE137 pKa = 3.99KK138 pKa = 10.22KK139 pKa = 10.25CLIWGEE145 pKa = 4.45CYY147 pKa = 10.37CYY149 pKa = 10.05RR150 pKa = 11.84CYY152 pKa = 10.92LLWFGFPPTWEE163 pKa = 4.04TFDD166 pKa = 3.39WYY168 pKa = 11.13QEE170 pKa = 4.31IILNTDD176 pKa = 2.81MHH178 pKa = 7.17LLRR181 pKa = 11.84LFNYY185 pKa = 10.02CKK187 pKa = 9.85YY188 pKa = 10.57QPGGDD193 pKa = 3.21SHH195 pKa = 7.79QPGKK199 pKa = 10.83LCC201 pKa = 5.09
MM1 pKa = 7.99DD2 pKa = 6.15SILSRR7 pKa = 11.84QEE9 pKa = 3.63KK10 pKa = 10.49RR11 pKa = 11.84EE12 pKa = 3.98LCDD15 pKa = 3.54LLEE18 pKa = 4.82IPHH21 pKa = 6.66HH22 pKa = 6.83CYY24 pKa = 11.18GNIPMMKK31 pKa = 9.43AQYY34 pKa = 11.28KK35 pKa = 9.96KK36 pKa = 9.57MCLLYY41 pKa = 10.77HH42 pKa = 6.92PDD44 pKa = 3.64KK45 pKa = 11.46GGDD48 pKa = 3.38GAKK51 pKa = 9.02MVKK54 pKa = 10.2LNSLWTSFQDD64 pKa = 3.28EE65 pKa = 4.63VTKK68 pKa = 10.92LRR70 pKa = 11.84AEE72 pKa = 4.3VNFFSYY78 pKa = 10.14QVSAKK83 pKa = 9.8FFWDD87 pKa = 3.45LDD89 pKa = 3.87FNTLKK94 pKa = 10.68EE95 pKa = 4.18YY96 pKa = 10.72LGKK99 pKa = 9.76NQCKK103 pKa = 9.56FLRR106 pKa = 11.84GPNCLNSKK114 pKa = 10.22YY115 pKa = 10.34SYY117 pKa = 10.1CRR119 pKa = 11.84CIVCRR124 pKa = 11.84LSQQHH129 pKa = 5.93QDD131 pKa = 3.07IKK133 pKa = 10.81DD134 pKa = 3.75TQEE137 pKa = 3.99KK138 pKa = 10.22KK139 pKa = 10.25CLIWGEE145 pKa = 4.45CYY147 pKa = 10.37CYY149 pKa = 10.05RR150 pKa = 11.84CYY152 pKa = 10.92LLWFGFPPTWEE163 pKa = 4.04TFDD166 pKa = 3.39WYY168 pKa = 11.13QEE170 pKa = 4.31IILNTDD176 pKa = 2.81MHH178 pKa = 7.17LLRR181 pKa = 11.84LFNYY185 pKa = 10.02CKK187 pKa = 9.85YY188 pKa = 10.57QPGGDD193 pKa = 3.21SHH195 pKa = 7.79QPGKK199 pKa = 10.83LCC201 pKa = 5.09
Molecular weight: 23.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2014 |
193 |
781 |
335.7 |
37.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.703 ± 1.144 | 2.334 ± 0.577 |
4.667 ± 0.397 | 6.504 ± 0.46 |
4.171 ± 0.61 | 6.703 ± 0.919 |
2.135 ± 0.519 | 5.065 ± 0.451 |
6.455 ± 1.082 | 9.732 ± 0.51 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.936 ± 0.406 | 4.27 ± 0.242 |
6.604 ± 1.54 | 4.568 ± 0.648 |
4.518 ± 0.601 | 8.193 ± 1.173 |
5.76 ± 0.342 | 4.965 ± 0.675 |
1.44 ± 0.318 | 3.277 ± 0.662 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
