
Epizootic hemorrhagic disease virus (serotype 1 / strain New Jersey)
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus; Epizootic hemorrhagic disease virus; Epizootic hemorrhagic disease virus 1
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
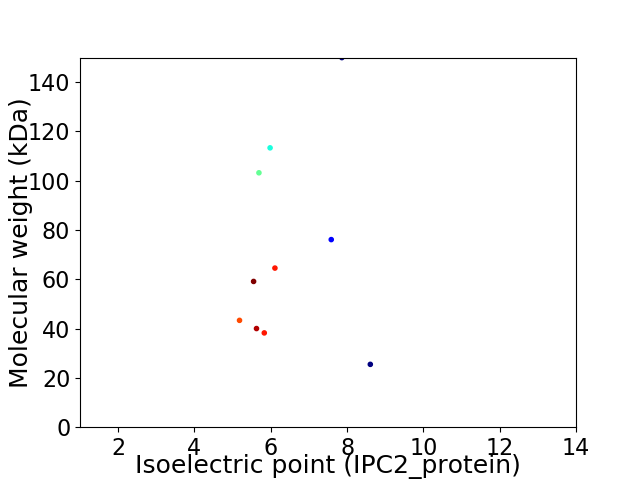
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
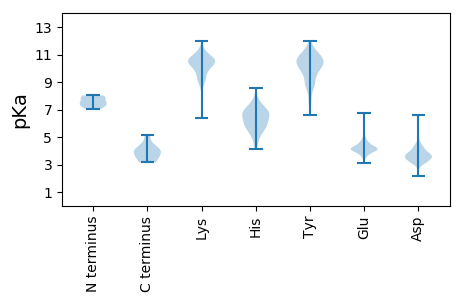
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C8TE66|C8TE66_EHDV1 VP6 protein OS=Epizootic hemorrhagic disease virus (serotype 1 / strain New Jersey) OX=449133 GN=VP6 PE=4 SV=1
MM1 pKa = 7.04EE2 pKa = 4.1QKK4 pKa = 9.85QRR6 pKa = 11.84RR7 pKa = 11.84FTKK10 pKa = 10.4NVFVLDD16 pKa = 3.85QKK18 pKa = 11.27RR19 pKa = 11.84KK20 pKa = 6.48TLCGQIAARR29 pKa = 11.84QSLPYY34 pKa = 9.69CQIKK38 pKa = 9.85IGRR41 pKa = 11.84NFALKK46 pKa = 10.27AVATPEE52 pKa = 3.8PKK54 pKa = 10.48GYY56 pKa = 10.52VLEE59 pKa = 4.56ICDD62 pKa = 3.36VGAYY66 pKa = 10.01RR67 pKa = 11.84IQDD70 pKa = 3.68GNDD73 pKa = 3.35VISLMISADD82 pKa = 3.52GVEE85 pKa = 4.75GTQEE89 pKa = 3.66RR90 pKa = 11.84WEE92 pKa = 3.97EE93 pKa = 4.01WKK95 pKa = 10.86FEE97 pKa = 4.43TISCVPMATVLNINGALVDD116 pKa = 3.95AEE118 pKa = 4.3IKK120 pKa = 10.02VSKK123 pKa = 10.08GMGMVPPYY131 pKa = 9.61TRR133 pKa = 11.84NDD135 pKa = 3.09FDD137 pKa = 4.59RR138 pKa = 11.84RR139 pKa = 11.84EE140 pKa = 4.08LPEE143 pKa = 4.19LPGVQRR149 pKa = 11.84SKK151 pKa = 11.24YY152 pKa = 9.96DD153 pKa = 3.29IRR155 pKa = 11.84EE156 pKa = 3.83LRR158 pKa = 11.84QKK160 pKa = 10.21IRR162 pKa = 11.84EE163 pKa = 4.03EE164 pKa = 4.1RR165 pKa = 11.84EE166 pKa = 3.69KK167 pKa = 11.24GAVEE171 pKa = 4.21QPHH174 pKa = 6.38KK175 pKa = 10.35PAFKK179 pKa = 9.28TEE181 pKa = 3.91RR182 pKa = 11.84WHH184 pKa = 5.9EE185 pKa = 4.08QPDD188 pKa = 3.57SDD190 pKa = 4.94EE191 pKa = 4.56DD192 pKa = 3.54QNLAGGVVNDD202 pKa = 4.25WTCEE206 pKa = 3.94TQEE209 pKa = 4.01RR210 pKa = 11.84DD211 pKa = 3.12QEE213 pKa = 4.26AEE215 pKa = 3.69RR216 pKa = 11.84RR217 pKa = 11.84EE218 pKa = 3.97ALEE221 pKa = 3.68IRR223 pKa = 11.84LAEE226 pKa = 3.68QRR228 pKa = 11.84QRR230 pKa = 11.84IEE232 pKa = 3.79AFKK235 pKa = 10.79LDD237 pKa = 3.99SEE239 pKa = 4.63RR240 pKa = 11.84KK241 pKa = 9.28RR242 pKa = 11.84EE243 pKa = 3.92DD244 pKa = 3.2LEE246 pKa = 4.51RR247 pKa = 11.84SQEE250 pKa = 3.85QFRR253 pKa = 11.84TQEE256 pKa = 4.2VEE258 pKa = 3.72WKK260 pKa = 9.53KK261 pKa = 10.77PDD263 pKa = 3.18MGEE266 pKa = 4.2IIIEE270 pKa = 4.12EE271 pKa = 4.2DD272 pKa = 3.78DD273 pKa = 3.66EE274 pKa = 5.99DD275 pKa = 4.86SEE277 pKa = 4.56EE278 pKa = 4.33EE279 pKa = 3.82EE280 pKa = 4.55GARR283 pKa = 11.84ASYY286 pKa = 8.49ITSAYY291 pKa = 8.91IEE293 pKa = 4.62RR294 pKa = 11.84ISRR297 pKa = 11.84IRR299 pKa = 11.84KK300 pKa = 9.21IKK302 pKa = 10.19DD303 pKa = 2.93EE304 pKa = 4.35RR305 pKa = 11.84LSMLASMMPQQSGEE319 pKa = 4.06YY320 pKa = 6.62TTTLFIKK327 pKa = 9.15KK328 pKa = 9.36QKK330 pKa = 8.54WDD332 pKa = 3.81NVPLYY337 pKa = 10.77LIDD340 pKa = 4.24EE341 pKa = 4.34MQKK344 pKa = 10.31KK345 pKa = 9.72YY346 pKa = 10.64EE347 pKa = 4.01LQSVGSCEE355 pKa = 3.58RR356 pKa = 11.84VAFVSKK362 pKa = 9.14GTNLIILPVGVV373 pKa = 3.86
MM1 pKa = 7.04EE2 pKa = 4.1QKK4 pKa = 9.85QRR6 pKa = 11.84RR7 pKa = 11.84FTKK10 pKa = 10.4NVFVLDD16 pKa = 3.85QKK18 pKa = 11.27RR19 pKa = 11.84KK20 pKa = 6.48TLCGQIAARR29 pKa = 11.84QSLPYY34 pKa = 9.69CQIKK38 pKa = 9.85IGRR41 pKa = 11.84NFALKK46 pKa = 10.27AVATPEE52 pKa = 3.8PKK54 pKa = 10.48GYY56 pKa = 10.52VLEE59 pKa = 4.56ICDD62 pKa = 3.36VGAYY66 pKa = 10.01RR67 pKa = 11.84IQDD70 pKa = 3.68GNDD73 pKa = 3.35VISLMISADD82 pKa = 3.52GVEE85 pKa = 4.75GTQEE89 pKa = 3.66RR90 pKa = 11.84WEE92 pKa = 3.97EE93 pKa = 4.01WKK95 pKa = 10.86FEE97 pKa = 4.43TISCVPMATVLNINGALVDD116 pKa = 3.95AEE118 pKa = 4.3IKK120 pKa = 10.02VSKK123 pKa = 10.08GMGMVPPYY131 pKa = 9.61TRR133 pKa = 11.84NDD135 pKa = 3.09FDD137 pKa = 4.59RR138 pKa = 11.84RR139 pKa = 11.84EE140 pKa = 4.08LPEE143 pKa = 4.19LPGVQRR149 pKa = 11.84SKK151 pKa = 11.24YY152 pKa = 9.96DD153 pKa = 3.29IRR155 pKa = 11.84EE156 pKa = 3.83LRR158 pKa = 11.84QKK160 pKa = 10.21IRR162 pKa = 11.84EE163 pKa = 4.03EE164 pKa = 4.1RR165 pKa = 11.84EE166 pKa = 3.69KK167 pKa = 11.24GAVEE171 pKa = 4.21QPHH174 pKa = 6.38KK175 pKa = 10.35PAFKK179 pKa = 9.28TEE181 pKa = 3.91RR182 pKa = 11.84WHH184 pKa = 5.9EE185 pKa = 4.08QPDD188 pKa = 3.57SDD190 pKa = 4.94EE191 pKa = 4.56DD192 pKa = 3.54QNLAGGVVNDD202 pKa = 4.25WTCEE206 pKa = 3.94TQEE209 pKa = 4.01RR210 pKa = 11.84DD211 pKa = 3.12QEE213 pKa = 4.26AEE215 pKa = 3.69RR216 pKa = 11.84RR217 pKa = 11.84EE218 pKa = 3.97ALEE221 pKa = 3.68IRR223 pKa = 11.84LAEE226 pKa = 3.68QRR228 pKa = 11.84QRR230 pKa = 11.84IEE232 pKa = 3.79AFKK235 pKa = 10.79LDD237 pKa = 3.99SEE239 pKa = 4.63RR240 pKa = 11.84KK241 pKa = 9.28RR242 pKa = 11.84EE243 pKa = 3.92DD244 pKa = 3.2LEE246 pKa = 4.51RR247 pKa = 11.84SQEE250 pKa = 3.85QFRR253 pKa = 11.84TQEE256 pKa = 4.2VEE258 pKa = 3.72WKK260 pKa = 9.53KK261 pKa = 10.77PDD263 pKa = 3.18MGEE266 pKa = 4.2IIIEE270 pKa = 4.12EE271 pKa = 4.2DD272 pKa = 3.78DD273 pKa = 3.66EE274 pKa = 5.99DD275 pKa = 4.86SEE277 pKa = 4.56EE278 pKa = 4.33EE279 pKa = 3.82EE280 pKa = 4.55GARR283 pKa = 11.84ASYY286 pKa = 8.49ITSAYY291 pKa = 8.91IEE293 pKa = 4.62RR294 pKa = 11.84ISRR297 pKa = 11.84IRR299 pKa = 11.84KK300 pKa = 9.21IKK302 pKa = 10.19DD303 pKa = 2.93EE304 pKa = 4.35RR305 pKa = 11.84LSMLASMMPQQSGEE319 pKa = 4.06YY320 pKa = 6.62TTTLFIKK327 pKa = 9.15KK328 pKa = 9.36QKK330 pKa = 8.54WDD332 pKa = 3.81NVPLYY337 pKa = 10.77LIDD340 pKa = 4.24EE341 pKa = 4.34MQKK344 pKa = 10.31KK345 pKa = 9.72YY346 pKa = 10.64EE347 pKa = 4.01LQSVGSCEE355 pKa = 3.58RR356 pKa = 11.84VAFVSKK362 pKa = 9.14GTNLIILPVGVV373 pKa = 3.86
Molecular weight: 43.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C9QNK7|C9QNK7_EHDV1 Non-structural protein NS3 OS=Epizootic hemorrhagic disease virus (serotype 1 / strain New Jersey) OX=449133 GN=NS3 PE=3 SV=1
MM1 pKa = 7.88LSRR4 pKa = 11.84LVPGVEE10 pKa = 4.01TRR12 pKa = 11.84IEE14 pKa = 4.02MKK16 pKa = 10.77ASDD19 pKa = 4.27EE20 pKa = 4.26VSLVPYY26 pKa = 9.71QEE28 pKa = 4.0QVRR31 pKa = 11.84PPSYY35 pKa = 10.68VPSAPIPTAMPRR47 pKa = 11.84VALDD51 pKa = 3.46ILDD54 pKa = 3.97KK55 pKa = 11.5AMSNQTGATMAQKK68 pKa = 10.38VEE70 pKa = 4.11KK71 pKa = 10.19VAYY74 pKa = 9.76ASYY77 pKa = 11.54AEE79 pKa = 4.37AFRR82 pKa = 11.84DD83 pKa = 3.96DD84 pKa = 3.69LRR86 pKa = 11.84LRR88 pKa = 11.84QIKK91 pKa = 9.97KK92 pKa = 9.74HH93 pKa = 5.3VNEE96 pKa = 4.13QILPKK101 pKa = 9.92MRR103 pKa = 11.84VEE105 pKa = 4.12LTAMKK110 pKa = 10.09RR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84AMAHH117 pKa = 5.06MVLIIAAVVALITSASTLTSDD138 pKa = 4.25LGVILKK144 pKa = 10.67HH145 pKa = 5.3NTTTEE150 pKa = 4.75AIQTYY155 pKa = 10.06IKK157 pKa = 9.49PFCAAFGIINLAATMVMMFMAKK179 pKa = 9.51NEE181 pKa = 4.26KK182 pKa = 10.04IINQQIDD189 pKa = 3.54HH190 pKa = 5.65TRR192 pKa = 11.84KK193 pKa = 10.24EE194 pKa = 4.06IMKK197 pKa = 9.93KK198 pKa = 10.03DD199 pKa = 3.6AYY201 pKa = 10.75NEE203 pKa = 3.94AVRR206 pKa = 11.84MSVTEE211 pKa = 3.98FSGIPLDD218 pKa = 4.54GFDD221 pKa = 5.09IPPEE225 pKa = 4.01LTRR228 pKa = 5.13
MM1 pKa = 7.88LSRR4 pKa = 11.84LVPGVEE10 pKa = 4.01TRR12 pKa = 11.84IEE14 pKa = 4.02MKK16 pKa = 10.77ASDD19 pKa = 4.27EE20 pKa = 4.26VSLVPYY26 pKa = 9.71QEE28 pKa = 4.0QVRR31 pKa = 11.84PPSYY35 pKa = 10.68VPSAPIPTAMPRR47 pKa = 11.84VALDD51 pKa = 3.46ILDD54 pKa = 3.97KK55 pKa = 11.5AMSNQTGATMAQKK68 pKa = 10.38VEE70 pKa = 4.11KK71 pKa = 10.19VAYY74 pKa = 9.76ASYY77 pKa = 11.54AEE79 pKa = 4.37AFRR82 pKa = 11.84DD83 pKa = 3.96DD84 pKa = 3.69LRR86 pKa = 11.84LRR88 pKa = 11.84QIKK91 pKa = 9.97KK92 pKa = 9.74HH93 pKa = 5.3VNEE96 pKa = 4.13QILPKK101 pKa = 9.92MRR103 pKa = 11.84VEE105 pKa = 4.12LTAMKK110 pKa = 10.09RR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84AMAHH117 pKa = 5.06MVLIIAAVVALITSASTLTSDD138 pKa = 4.25LGVILKK144 pKa = 10.67HH145 pKa = 5.3NTTTEE150 pKa = 4.75AIQTYY155 pKa = 10.06IKK157 pKa = 9.49PFCAAFGIINLAATMVMMFMAKK179 pKa = 9.51NEE181 pKa = 4.26KK182 pKa = 10.04IINQQIDD189 pKa = 3.54HH190 pKa = 5.65TRR192 pKa = 11.84KK193 pKa = 10.24EE194 pKa = 4.06IMKK197 pKa = 9.93KK198 pKa = 10.03DD199 pKa = 3.6AYY201 pKa = 10.75NEE203 pKa = 3.94AVRR206 pKa = 11.84MSVTEE211 pKa = 3.98FSGIPLDD218 pKa = 4.54GFDD221 pKa = 5.09IPPEE225 pKa = 4.01LTRR228 pKa = 5.13
Molecular weight: 25.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6204 |
228 |
1302 |
620.4 |
71.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.399 ± 0.556 | 1.177 ± 0.242 |
6.044 ± 0.364 | 7.963 ± 0.612 |
3.82 ± 0.302 | 5.77 ± 0.339 |
2.144 ± 0.234 | 7.447 ± 0.488 |
6.254 ± 0.684 | 8.333 ± 0.354 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.594 ± 0.422 | 3.965 ± 0.387 |
3.949 ± 0.305 | 4.094 ± 0.283 |
7.205 ± 0.385 | 5.255 ± 0.353 |
4.932 ± 0.322 | 6.593 ± 0.331 |
1.209 ± 0.242 | 3.852 ± 0.23 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
