
Glutamicibacter arilaitensis (strain DSM 16368 / CIP 108037 / IAM 15318 / JCM 13566 / NCIMB 14258 / Re117) (Arthrobacter arilaitensis)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Micrococcaceae; Glutamicibacter; Glutamicibacter arilaitensis
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
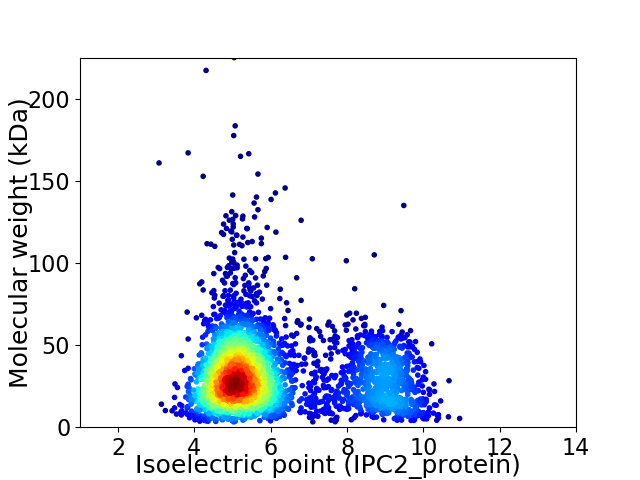
Virtual 2D-PAGE plot for 3380 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E1VT29|E1VT29_GLUAR ESAT-6-like protein OS=Glutamicibacter arilaitensis (strain DSM 16368 / CIP 108037 / IAM 15318 / JCM 13566 / NCIMB 14258 / Re117) OX=861360 GN=AARI_05540 PE=3 SV=1
MM1 pKa = 7.01MRR3 pKa = 11.84LAAVGTALAMGLAGCSSSTADD24 pKa = 4.01SQPEE28 pKa = 4.13DD29 pKa = 3.72AQSSQPAAKK38 pKa = 10.16SSAPEE43 pKa = 4.06GLEE46 pKa = 3.72SFYY49 pKa = 10.68TQEE52 pKa = 5.27LDD54 pKa = 3.07WEE56 pKa = 4.55MCGSIIEE63 pKa = 4.41CATIKK68 pKa = 10.88VPMDD72 pKa = 3.34YY73 pKa = 11.1ANPEE77 pKa = 4.02GQSIDD82 pKa = 3.31LALNRR87 pKa = 11.84RR88 pKa = 11.84AVDD91 pKa = 3.47GATGNILVNPGGPGGSGLDD110 pKa = 3.54MVSSTVPSMFSNDD123 pKa = 2.87LQRR126 pKa = 11.84AYY128 pKa = 11.01NVIGFDD134 pKa = 3.42PRR136 pKa = 11.84GVGEE140 pKa = 4.41SSPVTCQSAAEE151 pKa = 4.12TDD153 pKa = 3.09KK154 pKa = 11.33GRR156 pKa = 11.84QEE158 pKa = 4.1NLRR161 pKa = 11.84AWVPEE166 pKa = 3.99DD167 pKa = 2.97QHH169 pKa = 7.57EE170 pKa = 4.64IIEE173 pKa = 4.4STEE176 pKa = 4.36DD177 pKa = 3.49YY178 pKa = 11.29AADD181 pKa = 3.76CASNTGEE188 pKa = 4.77LLDD191 pKa = 4.89HH192 pKa = 6.93VDD194 pKa = 3.58TVSAARR200 pKa = 11.84DD201 pKa = 3.38MDD203 pKa = 4.24VIRR206 pKa = 11.84AVLGDD211 pKa = 3.7AQLDD215 pKa = 4.06YY216 pKa = 11.55LGVSYY221 pKa = 8.33GTFLGATYY229 pKa = 11.29ADD231 pKa = 5.03LFPQNVGRR239 pKa = 11.84FVLDD243 pKa = 3.54GAMDD247 pKa = 4.71PSLPAAEE254 pKa = 4.27VTLAQAVGFEE264 pKa = 4.43KK265 pKa = 10.4EE266 pKa = 3.6TDD268 pKa = 2.89AWLASCLEE276 pKa = 4.16SDD278 pKa = 3.9ACPFTGTVEE287 pKa = 3.98EE288 pKa = 5.1AKK290 pKa = 10.79VQLQQFFAQVEE301 pKa = 4.37NEE303 pKa = 4.16PMTASDD309 pKa = 3.67GRR311 pKa = 11.84VVPIIDD317 pKa = 3.9FVNGFVLPLYY327 pKa = 10.59DD328 pKa = 4.49DD329 pKa = 4.81SSWSYY334 pKa = 10.02LTDD337 pKa = 3.36AMNAAINDD345 pKa = 3.82ADD347 pKa = 3.39VDD349 pKa = 4.36MILGFADD356 pKa = 4.72LSADD360 pKa = 3.92RR361 pKa = 11.84QSDD364 pKa = 3.44GTYY367 pKa = 10.83ASNSSDD373 pKa = 3.02AFTAINCLDD382 pKa = 3.6RR383 pKa = 11.84PMDD386 pKa = 3.85SSAEE390 pKa = 4.13TMQDD394 pKa = 2.8EE395 pKa = 4.68ATEE398 pKa = 4.18LMRR401 pKa = 11.84MAPTLGKK408 pKa = 9.68YY409 pKa = 9.21LAYY412 pKa = 10.48GSITCEE418 pKa = 3.25AWDD421 pKa = 4.0YY422 pKa = 11.67DD423 pKa = 4.0STGTPEE429 pKa = 4.46ILDD432 pKa = 3.69APGSNEE438 pKa = 3.39ILVIGTVGDD447 pKa = 3.48PATPYY452 pKa = 10.11KK453 pKa = 10.18WSEE456 pKa = 3.91ALAKK460 pKa = 10.55QLDD463 pKa = 3.86NATLLTYY470 pKa = 10.08EE471 pKa = 4.37GHH473 pKa = 6.0GHH475 pKa = 4.9TAYY478 pKa = 10.32GRR480 pKa = 11.84SNEE483 pKa = 4.64CITKK487 pKa = 10.54AVDD490 pKa = 3.29EE491 pKa = 4.2YY492 pKa = 11.39LIDD495 pKa = 4.11GKK497 pKa = 11.08VPEE500 pKa = 5.1AGTRR504 pKa = 11.84CC505 pKa = 3.49
MM1 pKa = 7.01MRR3 pKa = 11.84LAAVGTALAMGLAGCSSSTADD24 pKa = 4.01SQPEE28 pKa = 4.13DD29 pKa = 3.72AQSSQPAAKK38 pKa = 10.16SSAPEE43 pKa = 4.06GLEE46 pKa = 3.72SFYY49 pKa = 10.68TQEE52 pKa = 5.27LDD54 pKa = 3.07WEE56 pKa = 4.55MCGSIIEE63 pKa = 4.41CATIKK68 pKa = 10.88VPMDD72 pKa = 3.34YY73 pKa = 11.1ANPEE77 pKa = 4.02GQSIDD82 pKa = 3.31LALNRR87 pKa = 11.84RR88 pKa = 11.84AVDD91 pKa = 3.47GATGNILVNPGGPGGSGLDD110 pKa = 3.54MVSSTVPSMFSNDD123 pKa = 2.87LQRR126 pKa = 11.84AYY128 pKa = 11.01NVIGFDD134 pKa = 3.42PRR136 pKa = 11.84GVGEE140 pKa = 4.41SSPVTCQSAAEE151 pKa = 4.12TDD153 pKa = 3.09KK154 pKa = 11.33GRR156 pKa = 11.84QEE158 pKa = 4.1NLRR161 pKa = 11.84AWVPEE166 pKa = 3.99DD167 pKa = 2.97QHH169 pKa = 7.57EE170 pKa = 4.64IIEE173 pKa = 4.4STEE176 pKa = 4.36DD177 pKa = 3.49YY178 pKa = 11.29AADD181 pKa = 3.76CASNTGEE188 pKa = 4.77LLDD191 pKa = 4.89HH192 pKa = 6.93VDD194 pKa = 3.58TVSAARR200 pKa = 11.84DD201 pKa = 3.38MDD203 pKa = 4.24VIRR206 pKa = 11.84AVLGDD211 pKa = 3.7AQLDD215 pKa = 4.06YY216 pKa = 11.55LGVSYY221 pKa = 8.33GTFLGATYY229 pKa = 11.29ADD231 pKa = 5.03LFPQNVGRR239 pKa = 11.84FVLDD243 pKa = 3.54GAMDD247 pKa = 4.71PSLPAAEE254 pKa = 4.27VTLAQAVGFEE264 pKa = 4.43KK265 pKa = 10.4EE266 pKa = 3.6TDD268 pKa = 2.89AWLASCLEE276 pKa = 4.16SDD278 pKa = 3.9ACPFTGTVEE287 pKa = 3.98EE288 pKa = 5.1AKK290 pKa = 10.79VQLQQFFAQVEE301 pKa = 4.37NEE303 pKa = 4.16PMTASDD309 pKa = 3.67GRR311 pKa = 11.84VVPIIDD317 pKa = 3.9FVNGFVLPLYY327 pKa = 10.59DD328 pKa = 4.49DD329 pKa = 4.81SSWSYY334 pKa = 10.02LTDD337 pKa = 3.36AMNAAINDD345 pKa = 3.82ADD347 pKa = 3.39VDD349 pKa = 4.36MILGFADD356 pKa = 4.72LSADD360 pKa = 3.92RR361 pKa = 11.84QSDD364 pKa = 3.44GTYY367 pKa = 10.83ASNSSDD373 pKa = 3.02AFTAINCLDD382 pKa = 3.6RR383 pKa = 11.84PMDD386 pKa = 3.85SSAEE390 pKa = 4.13TMQDD394 pKa = 2.8EE395 pKa = 4.68ATEE398 pKa = 4.18LMRR401 pKa = 11.84MAPTLGKK408 pKa = 9.68YY409 pKa = 9.21LAYY412 pKa = 10.48GSITCEE418 pKa = 3.25AWDD421 pKa = 4.0YY422 pKa = 11.67DD423 pKa = 4.0STGTPEE429 pKa = 4.46ILDD432 pKa = 3.69APGSNEE438 pKa = 3.39ILVIGTVGDD447 pKa = 3.48PATPYY452 pKa = 10.11KK453 pKa = 10.18WSEE456 pKa = 3.91ALAKK460 pKa = 10.55QLDD463 pKa = 3.86NATLLTYY470 pKa = 10.08EE471 pKa = 4.37GHH473 pKa = 6.0GHH475 pKa = 4.9TAYY478 pKa = 10.32GRR480 pKa = 11.84SNEE483 pKa = 4.64CITKK487 pKa = 10.54AVDD490 pKa = 3.29EE491 pKa = 4.2YY492 pKa = 11.39LIDD495 pKa = 4.11GKK497 pKa = 11.08VPEE500 pKa = 5.1AGTRR504 pKa = 11.84CC505 pKa = 3.49
Molecular weight: 53.77 kDa
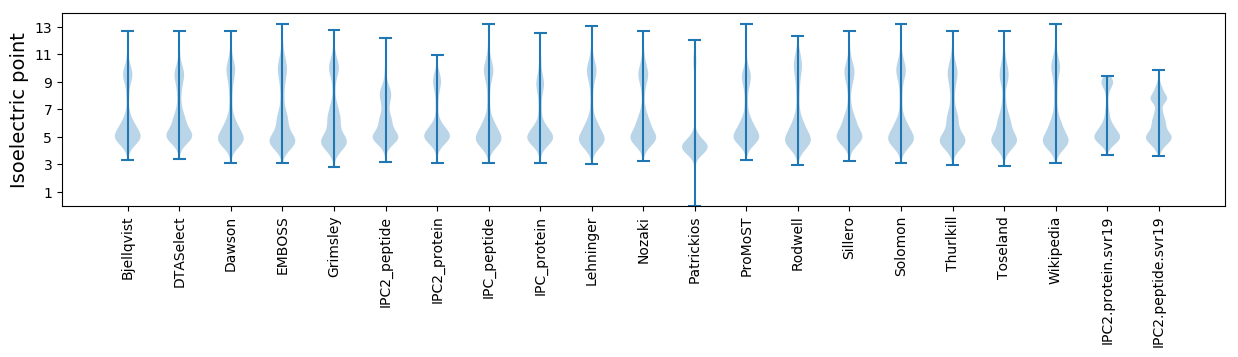
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E1VRE3|E1VRE3_GLUAR Uncharacterized protein OS=Glutamicibacter arilaitensis (strain DSM 16368 / CIP 108037 / IAM 15318 / JCM 13566 / NCIMB 14258 / Re117) OX=861360 GN=AARI_pI00410 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SKK15 pKa = 10.39KK16 pKa = 9.03HH17 pKa = 4.36GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSARR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 9.94NRR40 pKa = 11.84AEE42 pKa = 3.99LSAA45 pKa = 5.01
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SKK15 pKa = 10.39KK16 pKa = 9.03HH17 pKa = 4.36GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSARR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 9.94NRR40 pKa = 11.84AEE42 pKa = 3.99LSAA45 pKa = 5.01
Molecular weight: 5.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1062228 |
30 |
2173 |
314.3 |
34.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.065 ± 0.063 | 0.659 ± 0.011 |
5.42 ± 0.037 | 6.274 ± 0.041 |
3.317 ± 0.028 | 8.174 ± 0.036 |
2.175 ± 0.02 | 5.091 ± 0.029 |
3.516 ± 0.028 | 10.331 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.246 ± 0.017 | 2.89 ± 0.029 |
4.804 ± 0.032 | 3.782 ± 0.023 |
5.952 ± 0.048 | 6.343 ± 0.03 |
5.683 ± 0.034 | 7.68 ± 0.034 |
1.409 ± 0.019 | 2.19 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
