
Natronococcus occultus SP4
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Natrialbales; Natrialbaceae; Natronococcus; Natronococcus occultus
Average proteome isoelectric point is 4.82
Get precalculated fractions of proteins
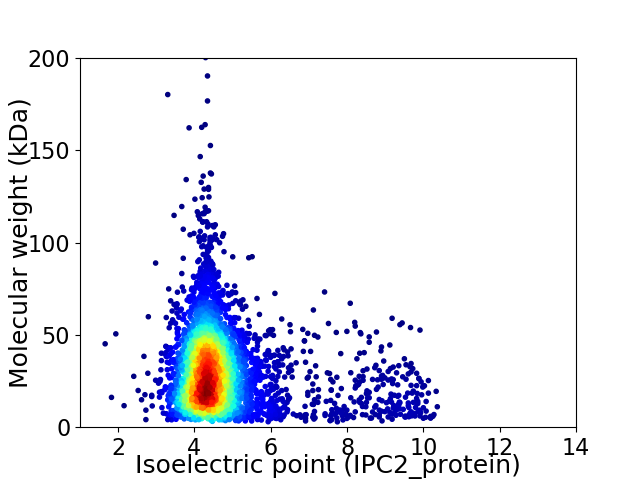
Virtual 2D-PAGE plot for 4153 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
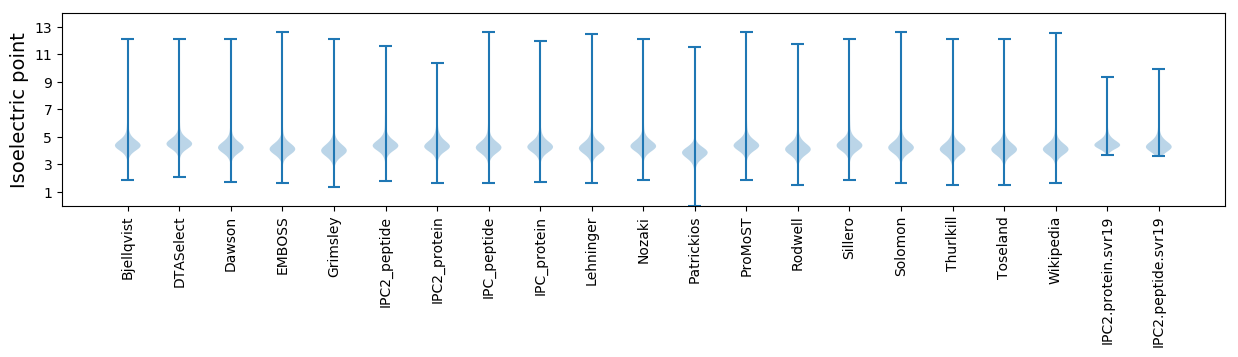
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L0K0A1|L0K0A1_9EURY Putative signal-transduction protein containing cAMP-binding and CBS domains OS=Natronococcus occultus SP4 OX=694430 GN=Natoc_1984 PE=4 SV=1
MM1 pKa = 6.69STEE4 pKa = 3.89TEE6 pKa = 3.9RR7 pKa = 11.84AEE9 pKa = 3.95EE10 pKa = 4.04RR11 pKa = 11.84TEE13 pKa = 4.24SSVLGSWAEE22 pKa = 4.12WYY24 pKa = 9.41HH25 pKa = 6.25VPVVGAVMLFMFWVRR40 pKa = 11.84TQSYY44 pKa = 9.04EE45 pKa = 4.1AFLTEE50 pKa = 5.42DD51 pKa = 3.74GTPALAGIDD60 pKa = 3.44SWYY63 pKa = 10.16HH64 pKa = 4.39WRR66 pKa = 11.84TIQWTAEE73 pKa = 4.09NYY75 pKa = 7.06PQTMPYY81 pKa = 9.7EE82 pKa = 4.28VWTGFPTGNYY92 pKa = 8.6VGQFGTLFDD101 pKa = 3.92QLIVTAAMIVGLGDD115 pKa = 3.72PSSSTLYY122 pKa = 9.84TVSLFAVPAMAALVAIPVFYY142 pKa = 10.72LGRR145 pKa = 11.84RR146 pKa = 11.84LGGTVGGLVAVVLLALAPGQFLVRR170 pKa = 11.84TTAGQLQHH178 pKa = 6.65HH179 pKa = 5.69VAEE182 pKa = 4.37VLFMALAVLGMAVALRR198 pKa = 11.84VAEE201 pKa = 4.12RR202 pKa = 11.84EE203 pKa = 3.94QPIYY207 pKa = 10.75EE208 pKa = 4.18LAADD212 pKa = 4.58RR213 pKa = 11.84NWAPLRR219 pKa = 11.84EE220 pKa = 4.08PTLYY224 pKa = 10.73SVLAGVALALYY235 pKa = 9.39IWVWPPGVVLIGIFGVFFAIQLCLDD260 pKa = 3.77YY261 pKa = 11.36VRR263 pKa = 11.84GISPDD268 pKa = 2.92HH269 pKa = 5.9VAFVGAVSLGVTAVATALLIEE290 pKa = 4.43EE291 pKa = 5.33PGTSVTSFGYY301 pKa = 9.58LQPGTAALVAVGCLFMAWFARR322 pKa = 11.84QWNARR327 pKa = 11.84GIDD330 pKa = 3.13RR331 pKa = 11.84RR332 pKa = 11.84LYY334 pKa = 9.61PGAIAALIVGTFAVMWLVLPEE355 pKa = 4.6FYY357 pKa = 9.4DD358 pKa = 3.7TLVGNLTGRR367 pKa = 11.84IIPLDD372 pKa = 4.03PDD374 pKa = 3.83AGTLTIQEE382 pKa = 4.32AQPPEE387 pKa = 4.97DD388 pKa = 3.59FTAHH392 pKa = 5.59VFDD395 pKa = 4.31EE396 pKa = 5.08FGTAFYY402 pKa = 10.89TMLAGLVLLAARR414 pKa = 11.84PLLGRR419 pKa = 11.84EE420 pKa = 3.73HH421 pKa = 6.65RR422 pKa = 11.84AEE424 pKa = 3.95YY425 pKa = 10.16TLILVWSLFLLSMSATQIRR444 pKa = 11.84FSYY447 pKa = 10.41YY448 pKa = 10.04LVLAVAVVNAAFVADD463 pKa = 3.76VLRR466 pKa = 11.84LFDD469 pKa = 5.85LDD471 pKa = 3.93LQRR474 pKa = 11.84GTQALRR480 pKa = 11.84GIEE483 pKa = 4.13TYY485 pKa = 10.58QIIILFVVVVLLFAPLLPVVAADD508 pKa = 3.43GDD510 pKa = 4.29TAVEE514 pKa = 4.18RR515 pKa = 11.84GEE517 pKa = 4.18ATGPHH522 pKa = 7.11PDD524 pKa = 3.05AMVWEE529 pKa = 5.15EE530 pKa = 4.06STHH533 pKa = 4.66WLEE536 pKa = 5.15ANTPEE541 pKa = 4.45PGNYY545 pKa = 9.75GGADD549 pKa = 3.31YY550 pKa = 11.25ADD552 pKa = 3.56EE553 pKa = 4.44LEE555 pKa = 4.65YY556 pKa = 11.07YY557 pKa = 10.65GSYY560 pKa = 10.5DD561 pKa = 3.61YY562 pKa = 11.45PEE564 pKa = 6.41DD565 pKa = 4.22GDD567 pKa = 3.71FDD569 pKa = 4.51YY570 pKa = 10.48PAGAYY575 pKa = 10.04GVMSWWDD582 pKa = 3.65YY583 pKa = 10.73GHH585 pKa = 7.77LITTQAEE592 pKa = 4.61RR593 pKa = 11.84IPHH596 pKa = 5.98SNPFQQNAEE605 pKa = 4.12TASAYY610 pKa = 8.42LTAEE614 pKa = 4.24SEE616 pKa = 4.44RR617 pKa = 11.84EE618 pKa = 3.85AEE620 pKa = 4.86LILEE624 pKa = 5.07AIAAGEE630 pKa = 4.56DD631 pKa = 4.01PLDD634 pKa = 4.7DD635 pKa = 5.55DD636 pKa = 5.97DD637 pKa = 4.79NVRR640 pKa = 11.84PLEE643 pKa = 4.04EE644 pKa = 5.45LEE646 pKa = 4.11TAIEE650 pKa = 4.47DD651 pKa = 4.72DD652 pKa = 3.73ADD654 pKa = 3.65EE655 pKa = 3.98QLRR658 pKa = 11.84YY659 pKa = 11.03VMIDD663 pKa = 3.2DD664 pKa = 3.69QMAGGKK670 pKa = 9.66FSAMTAWSGPDD681 pKa = 3.01YY682 pKa = 11.07DD683 pKa = 5.3HH684 pKa = 7.05YY685 pKa = 10.65VTPDD689 pKa = 3.35VEE691 pKa = 4.81PGEE694 pKa = 4.61SIDD697 pKa = 5.76RR698 pKa = 11.84EE699 pKa = 4.26DD700 pKa = 3.28VDD702 pKa = 5.78DD703 pKa = 4.3RR704 pKa = 11.84LKK706 pKa = 11.15VPYY709 pKa = 9.62EE710 pKa = 3.87EE711 pKa = 4.52TMVADD716 pKa = 5.51LYY718 pKa = 11.37FDD720 pKa = 4.88DD721 pKa = 4.32ATGMDD726 pKa = 4.47HH727 pKa = 6.81YY728 pKa = 11.32RR729 pKa = 11.84LVHH732 pKa = 6.24EE733 pKa = 4.53NDD735 pKa = 2.79EE736 pKa = 4.27RR737 pKa = 11.84TAGFVSYY744 pKa = 11.29ALIDD748 pKa = 3.99PEE750 pKa = 4.23TDD752 pKa = 2.91EE753 pKa = 4.89VLTDD757 pKa = 3.21EE758 pKa = 4.72TGQHH762 pKa = 4.76QVAFNQPLQQAEE774 pKa = 4.33VQLSQFEE781 pKa = 4.09QSPDD785 pKa = 3.0VDD787 pKa = 3.45VEE789 pKa = 4.36YY790 pKa = 10.6IDD792 pKa = 5.19DD793 pKa = 4.18RR794 pKa = 11.84EE795 pKa = 4.21TAGVKK800 pKa = 9.13TYY802 pKa = 10.74EE803 pKa = 4.18RR804 pKa = 11.84VAGATIVGTADD815 pKa = 3.21ADD817 pKa = 3.74ADD819 pKa = 3.87EE820 pKa = 5.38GDD822 pKa = 3.92VVTAEE827 pKa = 4.71LEE829 pKa = 4.25LDD831 pKa = 3.48SGTDD835 pKa = 2.97RR836 pKa = 11.84GSFTYY841 pKa = 10.09EE842 pKa = 3.56QEE844 pKa = 4.38TEE846 pKa = 3.65VDD848 pKa = 3.1ADD850 pKa = 4.14GEE852 pKa = 4.5FEE854 pKa = 4.41LTVPYY859 pKa = 9.72ATDD862 pKa = 3.91DD863 pKa = 3.78EE864 pKa = 6.09LDD866 pKa = 3.66VEE868 pKa = 4.8DD869 pKa = 6.14GYY871 pKa = 9.87TEE873 pKa = 4.24SAVEE877 pKa = 4.05AVDD880 pKa = 4.86DD881 pKa = 3.89YY882 pKa = 11.29TVTVEE887 pKa = 5.07GEE889 pKa = 4.32TPQEE893 pKa = 3.91GDD895 pKa = 3.67LEE897 pKa = 4.6VPEE900 pKa = 4.41SAVVDD905 pKa = 4.1GEE907 pKa = 4.55TVEE910 pKa = 4.58VEE912 pKa = 4.18LEE914 pKa = 4.01EE915 pKa = 4.63TEE917 pKa = 5.93LEE919 pKa = 4.19DD920 pKa = 5.47AEE922 pKa = 4.64NDD924 pKa = 3.68GATEE928 pKa = 4.65DD929 pKa = 4.03EE930 pKa = 4.53TDD932 pKa = 4.73GDD934 pKa = 4.1DD935 pKa = 4.72ADD937 pKa = 3.99AAEE940 pKa = 5.21ADD942 pKa = 4.17TEE944 pKa = 4.44TADD947 pKa = 4.89EE948 pKa = 4.85DD949 pKa = 5.04ADD951 pKa = 4.03DD952 pKa = 4.93AAANDD957 pKa = 3.73ADD959 pKa = 4.3AEE961 pKa = 4.32AGADD965 pKa = 3.97DD966 pKa = 5.59AGADD970 pKa = 3.84DD971 pKa = 4.57TASSLAPAAGLADD984 pKa = 3.56
MM1 pKa = 6.69STEE4 pKa = 3.89TEE6 pKa = 3.9RR7 pKa = 11.84AEE9 pKa = 3.95EE10 pKa = 4.04RR11 pKa = 11.84TEE13 pKa = 4.24SSVLGSWAEE22 pKa = 4.12WYY24 pKa = 9.41HH25 pKa = 6.25VPVVGAVMLFMFWVRR40 pKa = 11.84TQSYY44 pKa = 9.04EE45 pKa = 4.1AFLTEE50 pKa = 5.42DD51 pKa = 3.74GTPALAGIDD60 pKa = 3.44SWYY63 pKa = 10.16HH64 pKa = 4.39WRR66 pKa = 11.84TIQWTAEE73 pKa = 4.09NYY75 pKa = 7.06PQTMPYY81 pKa = 9.7EE82 pKa = 4.28VWTGFPTGNYY92 pKa = 8.6VGQFGTLFDD101 pKa = 3.92QLIVTAAMIVGLGDD115 pKa = 3.72PSSSTLYY122 pKa = 9.84TVSLFAVPAMAALVAIPVFYY142 pKa = 10.72LGRR145 pKa = 11.84RR146 pKa = 11.84LGGTVGGLVAVVLLALAPGQFLVRR170 pKa = 11.84TTAGQLQHH178 pKa = 6.65HH179 pKa = 5.69VAEE182 pKa = 4.37VLFMALAVLGMAVALRR198 pKa = 11.84VAEE201 pKa = 4.12RR202 pKa = 11.84EE203 pKa = 3.94QPIYY207 pKa = 10.75EE208 pKa = 4.18LAADD212 pKa = 4.58RR213 pKa = 11.84NWAPLRR219 pKa = 11.84EE220 pKa = 4.08PTLYY224 pKa = 10.73SVLAGVALALYY235 pKa = 9.39IWVWPPGVVLIGIFGVFFAIQLCLDD260 pKa = 3.77YY261 pKa = 11.36VRR263 pKa = 11.84GISPDD268 pKa = 2.92HH269 pKa = 5.9VAFVGAVSLGVTAVATALLIEE290 pKa = 4.43EE291 pKa = 5.33PGTSVTSFGYY301 pKa = 9.58LQPGTAALVAVGCLFMAWFARR322 pKa = 11.84QWNARR327 pKa = 11.84GIDD330 pKa = 3.13RR331 pKa = 11.84RR332 pKa = 11.84LYY334 pKa = 9.61PGAIAALIVGTFAVMWLVLPEE355 pKa = 4.6FYY357 pKa = 9.4DD358 pKa = 3.7TLVGNLTGRR367 pKa = 11.84IIPLDD372 pKa = 4.03PDD374 pKa = 3.83AGTLTIQEE382 pKa = 4.32AQPPEE387 pKa = 4.97DD388 pKa = 3.59FTAHH392 pKa = 5.59VFDD395 pKa = 4.31EE396 pKa = 5.08FGTAFYY402 pKa = 10.89TMLAGLVLLAARR414 pKa = 11.84PLLGRR419 pKa = 11.84EE420 pKa = 3.73HH421 pKa = 6.65RR422 pKa = 11.84AEE424 pKa = 3.95YY425 pKa = 10.16TLILVWSLFLLSMSATQIRR444 pKa = 11.84FSYY447 pKa = 10.41YY448 pKa = 10.04LVLAVAVVNAAFVADD463 pKa = 3.76VLRR466 pKa = 11.84LFDD469 pKa = 5.85LDD471 pKa = 3.93LQRR474 pKa = 11.84GTQALRR480 pKa = 11.84GIEE483 pKa = 4.13TYY485 pKa = 10.58QIIILFVVVVLLFAPLLPVVAADD508 pKa = 3.43GDD510 pKa = 4.29TAVEE514 pKa = 4.18RR515 pKa = 11.84GEE517 pKa = 4.18ATGPHH522 pKa = 7.11PDD524 pKa = 3.05AMVWEE529 pKa = 5.15EE530 pKa = 4.06STHH533 pKa = 4.66WLEE536 pKa = 5.15ANTPEE541 pKa = 4.45PGNYY545 pKa = 9.75GGADD549 pKa = 3.31YY550 pKa = 11.25ADD552 pKa = 3.56EE553 pKa = 4.44LEE555 pKa = 4.65YY556 pKa = 11.07YY557 pKa = 10.65GSYY560 pKa = 10.5DD561 pKa = 3.61YY562 pKa = 11.45PEE564 pKa = 6.41DD565 pKa = 4.22GDD567 pKa = 3.71FDD569 pKa = 4.51YY570 pKa = 10.48PAGAYY575 pKa = 10.04GVMSWWDD582 pKa = 3.65YY583 pKa = 10.73GHH585 pKa = 7.77LITTQAEE592 pKa = 4.61RR593 pKa = 11.84IPHH596 pKa = 5.98SNPFQQNAEE605 pKa = 4.12TASAYY610 pKa = 8.42LTAEE614 pKa = 4.24SEE616 pKa = 4.44RR617 pKa = 11.84EE618 pKa = 3.85AEE620 pKa = 4.86LILEE624 pKa = 5.07AIAAGEE630 pKa = 4.56DD631 pKa = 4.01PLDD634 pKa = 4.7DD635 pKa = 5.55DD636 pKa = 5.97DD637 pKa = 4.79NVRR640 pKa = 11.84PLEE643 pKa = 4.04EE644 pKa = 5.45LEE646 pKa = 4.11TAIEE650 pKa = 4.47DD651 pKa = 4.72DD652 pKa = 3.73ADD654 pKa = 3.65EE655 pKa = 3.98QLRR658 pKa = 11.84YY659 pKa = 11.03VMIDD663 pKa = 3.2DD664 pKa = 3.69QMAGGKK670 pKa = 9.66FSAMTAWSGPDD681 pKa = 3.01YY682 pKa = 11.07DD683 pKa = 5.3HH684 pKa = 7.05YY685 pKa = 10.65VTPDD689 pKa = 3.35VEE691 pKa = 4.81PGEE694 pKa = 4.61SIDD697 pKa = 5.76RR698 pKa = 11.84EE699 pKa = 4.26DD700 pKa = 3.28VDD702 pKa = 5.78DD703 pKa = 4.3RR704 pKa = 11.84LKK706 pKa = 11.15VPYY709 pKa = 9.62EE710 pKa = 3.87EE711 pKa = 4.52TMVADD716 pKa = 5.51LYY718 pKa = 11.37FDD720 pKa = 4.88DD721 pKa = 4.32ATGMDD726 pKa = 4.47HH727 pKa = 6.81YY728 pKa = 11.32RR729 pKa = 11.84LVHH732 pKa = 6.24EE733 pKa = 4.53NDD735 pKa = 2.79EE736 pKa = 4.27RR737 pKa = 11.84TAGFVSYY744 pKa = 11.29ALIDD748 pKa = 3.99PEE750 pKa = 4.23TDD752 pKa = 2.91EE753 pKa = 4.89VLTDD757 pKa = 3.21EE758 pKa = 4.72TGQHH762 pKa = 4.76QVAFNQPLQQAEE774 pKa = 4.33VQLSQFEE781 pKa = 4.09QSPDD785 pKa = 3.0VDD787 pKa = 3.45VEE789 pKa = 4.36YY790 pKa = 10.6IDD792 pKa = 5.19DD793 pKa = 4.18RR794 pKa = 11.84EE795 pKa = 4.21TAGVKK800 pKa = 9.13TYY802 pKa = 10.74EE803 pKa = 4.18RR804 pKa = 11.84VAGATIVGTADD815 pKa = 3.21ADD817 pKa = 3.74ADD819 pKa = 3.87EE820 pKa = 5.38GDD822 pKa = 3.92VVTAEE827 pKa = 4.71LEE829 pKa = 4.25LDD831 pKa = 3.48SGTDD835 pKa = 2.97RR836 pKa = 11.84GSFTYY841 pKa = 10.09EE842 pKa = 3.56QEE844 pKa = 4.38TEE846 pKa = 3.65VDD848 pKa = 3.1ADD850 pKa = 4.14GEE852 pKa = 4.5FEE854 pKa = 4.41LTVPYY859 pKa = 9.72ATDD862 pKa = 3.91DD863 pKa = 3.78EE864 pKa = 6.09LDD866 pKa = 3.66VEE868 pKa = 4.8DD869 pKa = 6.14GYY871 pKa = 9.87TEE873 pKa = 4.24SAVEE877 pKa = 4.05AVDD880 pKa = 4.86DD881 pKa = 3.89YY882 pKa = 11.29TVTVEE887 pKa = 5.07GEE889 pKa = 4.32TPQEE893 pKa = 3.91GDD895 pKa = 3.67LEE897 pKa = 4.6VPEE900 pKa = 4.41SAVVDD905 pKa = 4.1GEE907 pKa = 4.55TVEE910 pKa = 4.58VEE912 pKa = 4.18LEE914 pKa = 4.01EE915 pKa = 4.63TEE917 pKa = 5.93LEE919 pKa = 4.19DD920 pKa = 5.47AEE922 pKa = 4.64NDD924 pKa = 3.68GATEE928 pKa = 4.65DD929 pKa = 4.03EE930 pKa = 4.53TDD932 pKa = 4.73GDD934 pKa = 4.1DD935 pKa = 4.72ADD937 pKa = 3.99AAEE940 pKa = 5.21ADD942 pKa = 4.17TEE944 pKa = 4.44TADD947 pKa = 4.89EE948 pKa = 4.85DD949 pKa = 5.04ADD951 pKa = 4.03DD952 pKa = 4.93AAANDD957 pKa = 3.73ADD959 pKa = 4.3AEE961 pKa = 4.32AGADD965 pKa = 3.97DD966 pKa = 5.59AGADD970 pKa = 3.84DD971 pKa = 4.57TASSLAPAAGLADD984 pKa = 3.56
Molecular weight: 107.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L0JT47|L0JT47_9EURY NH(3)-dependent NAD(+) synthetase OS=Natronococcus occultus SP4 OX=694430 GN=nadE PE=3 SV=1
MM1 pKa = 7.38PVVSVSMPEE10 pKa = 3.81EE11 pKa = 3.67LLEE14 pKa = 4.6RR15 pKa = 11.84LDD17 pKa = 3.85AHH19 pKa = 6.91ASAHH23 pKa = 6.47EE24 pKa = 4.04YY25 pKa = 9.2TGRR28 pKa = 11.84SEE30 pKa = 4.46VVRR33 pKa = 11.84EE34 pKa = 4.0STRR37 pKa = 11.84ALLEE41 pKa = 4.07AFTDD45 pKa = 3.86DD46 pKa = 3.99RR47 pKa = 11.84VEE49 pKa = 4.72DD50 pKa = 3.89GALAGTVDD58 pKa = 2.98VRR60 pKa = 11.84YY61 pKa = 10.1DD62 pKa = 3.17VGTPSVEE69 pKa = 3.62RR70 pKa = 11.84RR71 pKa = 11.84VAALRR76 pKa = 11.84HH77 pKa = 5.49EE78 pKa = 4.49FDD80 pKa = 3.57EE81 pKa = 5.06AVAATDD87 pKa = 3.49HH88 pKa = 6.24SHH90 pKa = 7.0VGDD93 pKa = 3.82YY94 pKa = 11.24CPDD97 pKa = 3.74LFVLEE102 pKa = 4.73ADD104 pKa = 4.1RR105 pKa = 11.84EE106 pKa = 4.65TISSVVDD113 pKa = 2.88KK114 pKa = 10.52CGRR117 pKa = 11.84STEE120 pKa = 4.26PRR122 pKa = 11.84TSTTRR127 pKa = 11.84SSSSTADD134 pKa = 2.61RR135 pKa = 11.84VVHH138 pKa = 5.63SKK140 pKa = 10.42RR141 pKa = 11.84RR142 pKa = 11.84SHH144 pKa = 6.89PNAPIAARR152 pKa = 11.84RR153 pKa = 11.84GSVAGGKK160 pKa = 8.35VSRR163 pKa = 11.84LRR165 pKa = 11.84LLSLEE170 pKa = 4.3SRR172 pKa = 11.84WFEE175 pKa = 3.94CEE177 pKa = 3.47TRR179 pKa = 11.84SRR181 pKa = 11.84IPCHH185 pKa = 6.56RR186 pKa = 11.84PIPHH190 pKa = 6.91PPIPIRR196 pKa = 11.84SSPVRR201 pKa = 11.84PPQSGRR207 pKa = 11.84TVPRR211 pKa = 11.84TKK213 pKa = 10.4KK214 pKa = 10.07PSSTTSTVRR223 pKa = 11.84KK224 pKa = 9.23RR225 pKa = 11.84GSARR229 pKa = 11.84RR230 pKa = 11.84TPWRR234 pKa = 11.84SPTSNEE240 pKa = 3.94TGGSDD245 pKa = 3.29TASRR249 pKa = 11.84AQRR252 pKa = 11.84RR253 pKa = 11.84TSRR256 pKa = 11.84ISNPSRR262 pKa = 11.84VLSAWW267 pKa = 3.14
MM1 pKa = 7.38PVVSVSMPEE10 pKa = 3.81EE11 pKa = 3.67LLEE14 pKa = 4.6RR15 pKa = 11.84LDD17 pKa = 3.85AHH19 pKa = 6.91ASAHH23 pKa = 6.47EE24 pKa = 4.04YY25 pKa = 9.2TGRR28 pKa = 11.84SEE30 pKa = 4.46VVRR33 pKa = 11.84EE34 pKa = 4.0STRR37 pKa = 11.84ALLEE41 pKa = 4.07AFTDD45 pKa = 3.86DD46 pKa = 3.99RR47 pKa = 11.84VEE49 pKa = 4.72DD50 pKa = 3.89GALAGTVDD58 pKa = 2.98VRR60 pKa = 11.84YY61 pKa = 10.1DD62 pKa = 3.17VGTPSVEE69 pKa = 3.62RR70 pKa = 11.84RR71 pKa = 11.84VAALRR76 pKa = 11.84HH77 pKa = 5.49EE78 pKa = 4.49FDD80 pKa = 3.57EE81 pKa = 5.06AVAATDD87 pKa = 3.49HH88 pKa = 6.24SHH90 pKa = 7.0VGDD93 pKa = 3.82YY94 pKa = 11.24CPDD97 pKa = 3.74LFVLEE102 pKa = 4.73ADD104 pKa = 4.1RR105 pKa = 11.84EE106 pKa = 4.65TISSVVDD113 pKa = 2.88KK114 pKa = 10.52CGRR117 pKa = 11.84STEE120 pKa = 4.26PRR122 pKa = 11.84TSTTRR127 pKa = 11.84SSSSTADD134 pKa = 2.61RR135 pKa = 11.84VVHH138 pKa = 5.63SKK140 pKa = 10.42RR141 pKa = 11.84RR142 pKa = 11.84SHH144 pKa = 6.89PNAPIAARR152 pKa = 11.84RR153 pKa = 11.84GSVAGGKK160 pKa = 8.35VSRR163 pKa = 11.84LRR165 pKa = 11.84LLSLEE170 pKa = 4.3SRR172 pKa = 11.84WFEE175 pKa = 3.94CEE177 pKa = 3.47TRR179 pKa = 11.84SRR181 pKa = 11.84IPCHH185 pKa = 6.56RR186 pKa = 11.84PIPHH190 pKa = 6.91PPIPIRR196 pKa = 11.84SSPVRR201 pKa = 11.84PPQSGRR207 pKa = 11.84TVPRR211 pKa = 11.84TKK213 pKa = 10.4KK214 pKa = 10.07PSSTTSTVRR223 pKa = 11.84KK224 pKa = 9.23RR225 pKa = 11.84GSARR229 pKa = 11.84RR230 pKa = 11.84TPWRR234 pKa = 11.84SPTSNEE240 pKa = 3.94TGGSDD245 pKa = 3.29TASRR249 pKa = 11.84AQRR252 pKa = 11.84RR253 pKa = 11.84TSRR256 pKa = 11.84ISNPSRR262 pKa = 11.84VLSAWW267 pKa = 3.14
Molecular weight: 29.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1196774 |
29 |
1804 |
288.2 |
31.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.654 ± 0.057 | 0.765 ± 0.013 |
8.453 ± 0.054 | 9.608 ± 0.054 |
3.195 ± 0.027 | 8.314 ± 0.038 |
1.982 ± 0.017 | 4.288 ± 0.029 |
1.726 ± 0.026 | 9.13 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.697 ± 0.016 | 2.196 ± 0.018 |
4.651 ± 0.024 | 2.35 ± 0.022 |
6.656 ± 0.042 | 5.331 ± 0.03 |
6.262 ± 0.026 | 8.829 ± 0.037 |
1.151 ± 0.014 | 2.763 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
