
Synechococcus virus S-ESS1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
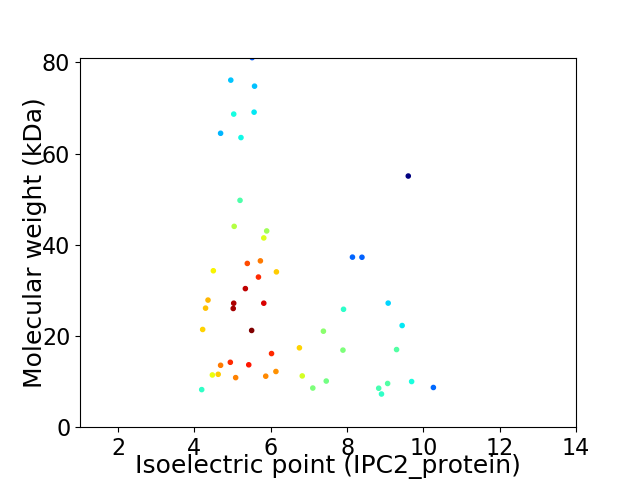
Virtual 2D-PAGE plot for 52 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1V0DX24|A0A1V0DX24_9CAUD Terminase large subunit OS=Synechococcus virus S-ESS1 OX=1964565 PE=3 SV=1
MM1 pKa = 7.38ARR3 pKa = 11.84NYY5 pKa = 9.69TLGRR9 pKa = 11.84GKK11 pKa = 10.19VHH13 pKa = 7.05FARR16 pKa = 11.84FKK18 pKa = 10.82PGTQVPDD25 pKa = 3.28GFFYY29 pKa = 10.49IGNTPEE35 pKa = 3.7FSLTIEE41 pKa = 4.43SEE43 pKa = 4.41SLDD46 pKa = 3.56HH47 pKa = 6.9FSSDD51 pKa = 2.72EE52 pKa = 4.36GIRR55 pKa = 11.84EE56 pKa = 4.0KK57 pKa = 11.01DD58 pKa = 3.24DD59 pKa = 3.95SVPLEE64 pKa = 4.02VNRR67 pKa = 11.84TGSLTTDD74 pKa = 3.51NIDD77 pKa = 3.73PEE79 pKa = 4.32NVALFFFGSSSVITQAAVASATEE102 pKa = 4.04TLTGIKK108 pKa = 10.01AGHH111 pKa = 5.9SYY113 pKa = 11.16KK114 pKa = 10.8LGVGPTNPAGYY125 pKa = 8.89MGIDD129 pKa = 3.22TTGFTVEE136 pKa = 4.15VGVTPLVVDD145 pKa = 3.65VDD147 pKa = 3.79YY148 pKa = 11.76TMDD151 pKa = 3.81FDD153 pKa = 4.27NGVITFLPDD162 pKa = 2.88SVLAVDD168 pKa = 4.2AADD171 pKa = 3.41VDD173 pKa = 3.94VTYY176 pKa = 11.07AVAASTRR183 pKa = 11.84SRR185 pKa = 11.84VISGSEE191 pKa = 3.83PVEE194 pKa = 3.83GAMMYY199 pKa = 8.36ITKK202 pKa = 10.19NPKK205 pKa = 8.79GTDD208 pKa = 3.14CTFYY212 pKa = 9.78MPYY215 pKa = 10.12VKK217 pKa = 10.55VMPNGDD223 pKa = 3.56YY224 pKa = 11.02ALKK227 pKa = 10.41GDD229 pKa = 4.09EE230 pKa = 4.11WQQIPLALEE239 pKa = 3.8VLKK242 pKa = 10.4PLTGEE247 pKa = 4.33AIYY250 pKa = 10.73RR251 pKa = 11.84DD252 pKa = 4.28GTPTMTPP259 pKa = 3.1
MM1 pKa = 7.38ARR3 pKa = 11.84NYY5 pKa = 9.69TLGRR9 pKa = 11.84GKK11 pKa = 10.19VHH13 pKa = 7.05FARR16 pKa = 11.84FKK18 pKa = 10.82PGTQVPDD25 pKa = 3.28GFFYY29 pKa = 10.49IGNTPEE35 pKa = 3.7FSLTIEE41 pKa = 4.43SEE43 pKa = 4.41SLDD46 pKa = 3.56HH47 pKa = 6.9FSSDD51 pKa = 2.72EE52 pKa = 4.36GIRR55 pKa = 11.84EE56 pKa = 4.0KK57 pKa = 11.01DD58 pKa = 3.24DD59 pKa = 3.95SVPLEE64 pKa = 4.02VNRR67 pKa = 11.84TGSLTTDD74 pKa = 3.51NIDD77 pKa = 3.73PEE79 pKa = 4.32NVALFFFGSSSVITQAAVASATEE102 pKa = 4.04TLTGIKK108 pKa = 10.01AGHH111 pKa = 5.9SYY113 pKa = 11.16KK114 pKa = 10.8LGVGPTNPAGYY125 pKa = 8.89MGIDD129 pKa = 3.22TTGFTVEE136 pKa = 4.15VGVTPLVVDD145 pKa = 3.65VDD147 pKa = 3.79YY148 pKa = 11.76TMDD151 pKa = 3.81FDD153 pKa = 4.27NGVITFLPDD162 pKa = 2.88SVLAVDD168 pKa = 4.2AADD171 pKa = 3.41VDD173 pKa = 3.94VTYY176 pKa = 11.07AVAASTRR183 pKa = 11.84SRR185 pKa = 11.84VISGSEE191 pKa = 3.83PVEE194 pKa = 3.83GAMMYY199 pKa = 8.36ITKK202 pKa = 10.19NPKK205 pKa = 8.79GTDD208 pKa = 3.14CTFYY212 pKa = 9.78MPYY215 pKa = 10.12VKK217 pKa = 10.55VMPNGDD223 pKa = 3.56YY224 pKa = 11.02ALKK227 pKa = 10.41GDD229 pKa = 4.09EE230 pKa = 4.11WQQIPLALEE239 pKa = 3.8VLKK242 pKa = 10.4PLTGEE247 pKa = 4.33AIYY250 pKa = 10.73RR251 pKa = 11.84DD252 pKa = 4.28GTPTMTPP259 pKa = 3.1
Molecular weight: 27.88 kDa
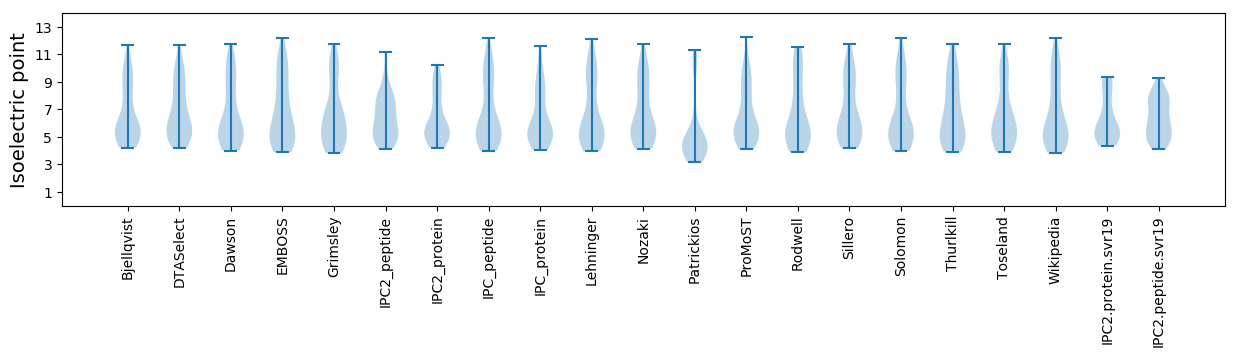
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V0DX53|A0A1V0DX53_9CAUD DUF3310 domain-containing protein OS=Synechococcus virus S-ESS1 OX=1964565 PE=4 SV=1
MM1 pKa = 6.99KK2 pKa = 9.86TVFSNHH8 pKa = 4.43MVAHH12 pKa = 5.9VFATQSQEE20 pKa = 3.7EE21 pKa = 4.59GRR23 pKa = 11.84SQNGNFYY30 pKa = 10.63FSGRR34 pKa = 11.84ALFSYY39 pKa = 10.06GRR41 pKa = 11.84HH42 pKa = 5.98FIVAYY47 pKa = 9.46AIPRR51 pKa = 11.84ISGGFLFLINPDD63 pKa = 3.54SYY65 pKa = 11.61SISTSRR71 pKa = 11.84HH72 pKa = 3.24QAMARR77 pKa = 11.84AAIPGRR83 pKa = 11.84AYY85 pKa = 9.78IAPNLTRR92 pKa = 11.84LADD95 pKa = 4.28DD96 pKa = 4.77LNAALDD102 pKa = 3.19AWEE105 pKa = 3.99YY106 pKa = 11.38HH107 pKa = 5.65EE108 pKa = 4.74MPGGRR113 pKa = 11.84TRR115 pKa = 11.84PVKK118 pKa = 10.4VAATMARR125 pKa = 11.84RR126 pKa = 11.84RR127 pKa = 11.84EE128 pKa = 3.83YY129 pKa = 10.78LPRR132 pKa = 11.84IKK134 pKa = 10.34RR135 pKa = 11.84AIAANWPGQEE145 pKa = 3.55TAAAILGEE153 pKa = 4.3FGQRR157 pKa = 11.84NAARR161 pKa = 11.84AAAAMARR168 pKa = 11.84RR169 pKa = 11.84HH170 pKa = 5.56AATVAAEE177 pKa = 4.14AAAKK181 pKa = 10.17AKK183 pKa = 10.33RR184 pKa = 11.84KK185 pKa = 10.18RR186 pKa = 11.84EE187 pKa = 3.92DD188 pKa = 2.92VARR191 pKa = 11.84NAKK194 pKa = 9.71YY195 pKa = 10.31LATFTPARR203 pKa = 11.84IRR205 pKa = 11.84AEE207 pKa = 3.47ISDD210 pKa = 3.58NLNRR214 pKa = 11.84YY215 pKa = 8.18SGQWGRR221 pKa = 11.84DD222 pKa = 3.04KK223 pKa = 10.98VKK225 pKa = 10.45EE226 pKa = 4.02MSRR229 pKa = 11.84HH230 pKa = 4.83LFAAIKK236 pKa = 9.14EE237 pKa = 4.04ARR239 pKa = 11.84ARR241 pKa = 11.84GWTRR245 pKa = 11.84IAATLSSHH253 pKa = 6.38RR254 pKa = 11.84AMVRR258 pKa = 11.84AALADD263 pKa = 3.38IDD265 pKa = 4.15NRR267 pKa = 11.84EE268 pKa = 3.79AMARR272 pKa = 11.84RR273 pKa = 11.84QGAKK277 pKa = 9.55RR278 pKa = 11.84EE279 pKa = 4.18AIARR283 pKa = 11.84FRR285 pKa = 11.84EE286 pKa = 4.12DD287 pKa = 2.89AANVARR293 pKa = 11.84LMTPRR298 pKa = 11.84DD299 pKa = 3.32AGGIAGEE306 pKa = 4.42GSPEE310 pKa = 3.76TRR312 pKa = 11.84ARR314 pKa = 11.84WYY316 pKa = 10.61RR317 pKa = 11.84DD318 pKa = 3.17LADD321 pKa = 3.88SCHH324 pKa = 6.32SLLRR328 pKa = 11.84HH329 pKa = 5.24WTPPAATVARR339 pKa = 11.84LRR341 pKa = 11.84SIHH344 pKa = 6.35DD345 pKa = 3.45AARR348 pKa = 11.84IRR350 pKa = 11.84MTEE353 pKa = 3.88EE354 pKa = 3.47NEE356 pKa = 3.59NAARR360 pKa = 11.84ARR362 pKa = 11.84FEE364 pKa = 4.34AEE366 pKa = 3.64AEE368 pKa = 4.04RR369 pKa = 11.84RR370 pKa = 11.84RR371 pKa = 11.84AWLAGEE377 pKa = 4.18TGLGYY382 pKa = 10.96GRR384 pKa = 11.84LSDD387 pKa = 3.73QDD389 pKa = 3.29GGALLRR395 pKa = 11.84AVDD398 pKa = 3.73VARR401 pKa = 11.84DD402 pKa = 3.42EE403 pKa = 4.56SGAIVSGVLEE413 pKa = 4.52TGHH416 pKa = 6.46GAQVPLVHH424 pKa = 7.19ALRR427 pKa = 11.84VFAFLRR433 pKa = 11.84HH434 pKa = 5.45CRR436 pKa = 11.84ATGRR440 pKa = 11.84AWKK443 pKa = 10.44RR444 pKa = 11.84NGASLRR450 pKa = 11.84VGHH453 pKa = 5.91FHH455 pKa = 6.87VDD457 pKa = 3.07SVTPQGDD464 pKa = 4.08FVAGCHH470 pKa = 6.18RR471 pKa = 11.84INWQEE476 pKa = 3.85VARR479 pKa = 11.84LAEE482 pKa = 4.14SLGVAEE488 pKa = 5.35LAPADD493 pKa = 3.98TTEE496 pKa = 3.91GHH498 pKa = 7.34AIAA501 pKa = 5.47
MM1 pKa = 6.99KK2 pKa = 9.86TVFSNHH8 pKa = 4.43MVAHH12 pKa = 5.9VFATQSQEE20 pKa = 3.7EE21 pKa = 4.59GRR23 pKa = 11.84SQNGNFYY30 pKa = 10.63FSGRR34 pKa = 11.84ALFSYY39 pKa = 10.06GRR41 pKa = 11.84HH42 pKa = 5.98FIVAYY47 pKa = 9.46AIPRR51 pKa = 11.84ISGGFLFLINPDD63 pKa = 3.54SYY65 pKa = 11.61SISTSRR71 pKa = 11.84HH72 pKa = 3.24QAMARR77 pKa = 11.84AAIPGRR83 pKa = 11.84AYY85 pKa = 9.78IAPNLTRR92 pKa = 11.84LADD95 pKa = 4.28DD96 pKa = 4.77LNAALDD102 pKa = 3.19AWEE105 pKa = 3.99YY106 pKa = 11.38HH107 pKa = 5.65EE108 pKa = 4.74MPGGRR113 pKa = 11.84TRR115 pKa = 11.84PVKK118 pKa = 10.4VAATMARR125 pKa = 11.84RR126 pKa = 11.84RR127 pKa = 11.84EE128 pKa = 3.83YY129 pKa = 10.78LPRR132 pKa = 11.84IKK134 pKa = 10.34RR135 pKa = 11.84AIAANWPGQEE145 pKa = 3.55TAAAILGEE153 pKa = 4.3FGQRR157 pKa = 11.84NAARR161 pKa = 11.84AAAAMARR168 pKa = 11.84RR169 pKa = 11.84HH170 pKa = 5.56AATVAAEE177 pKa = 4.14AAAKK181 pKa = 10.17AKK183 pKa = 10.33RR184 pKa = 11.84KK185 pKa = 10.18RR186 pKa = 11.84EE187 pKa = 3.92DD188 pKa = 2.92VARR191 pKa = 11.84NAKK194 pKa = 9.71YY195 pKa = 10.31LATFTPARR203 pKa = 11.84IRR205 pKa = 11.84AEE207 pKa = 3.47ISDD210 pKa = 3.58NLNRR214 pKa = 11.84YY215 pKa = 8.18SGQWGRR221 pKa = 11.84DD222 pKa = 3.04KK223 pKa = 10.98VKK225 pKa = 10.45EE226 pKa = 4.02MSRR229 pKa = 11.84HH230 pKa = 4.83LFAAIKK236 pKa = 9.14EE237 pKa = 4.04ARR239 pKa = 11.84ARR241 pKa = 11.84GWTRR245 pKa = 11.84IAATLSSHH253 pKa = 6.38RR254 pKa = 11.84AMVRR258 pKa = 11.84AALADD263 pKa = 3.38IDD265 pKa = 4.15NRR267 pKa = 11.84EE268 pKa = 3.79AMARR272 pKa = 11.84RR273 pKa = 11.84QGAKK277 pKa = 9.55RR278 pKa = 11.84EE279 pKa = 4.18AIARR283 pKa = 11.84FRR285 pKa = 11.84EE286 pKa = 4.12DD287 pKa = 2.89AANVARR293 pKa = 11.84LMTPRR298 pKa = 11.84DD299 pKa = 3.32AGGIAGEE306 pKa = 4.42GSPEE310 pKa = 3.76TRR312 pKa = 11.84ARR314 pKa = 11.84WYY316 pKa = 10.61RR317 pKa = 11.84DD318 pKa = 3.17LADD321 pKa = 3.88SCHH324 pKa = 6.32SLLRR328 pKa = 11.84HH329 pKa = 5.24WTPPAATVARR339 pKa = 11.84LRR341 pKa = 11.84SIHH344 pKa = 6.35DD345 pKa = 3.45AARR348 pKa = 11.84IRR350 pKa = 11.84MTEE353 pKa = 3.88EE354 pKa = 3.47NEE356 pKa = 3.59NAARR360 pKa = 11.84ARR362 pKa = 11.84FEE364 pKa = 4.34AEE366 pKa = 3.64AEE368 pKa = 4.04RR369 pKa = 11.84RR370 pKa = 11.84RR371 pKa = 11.84AWLAGEE377 pKa = 4.18TGLGYY382 pKa = 10.96GRR384 pKa = 11.84LSDD387 pKa = 3.73QDD389 pKa = 3.29GGALLRR395 pKa = 11.84AVDD398 pKa = 3.73VARR401 pKa = 11.84DD402 pKa = 3.42EE403 pKa = 4.56SGAIVSGVLEE413 pKa = 4.52TGHH416 pKa = 6.46GAQVPLVHH424 pKa = 7.19ALRR427 pKa = 11.84VFAFLRR433 pKa = 11.84HH434 pKa = 5.45CRR436 pKa = 11.84ATGRR440 pKa = 11.84AWKK443 pKa = 10.44RR444 pKa = 11.84NGASLRR450 pKa = 11.84VGHH453 pKa = 5.91FHH455 pKa = 6.87VDD457 pKa = 3.07SVTPQGDD464 pKa = 4.08FVAGCHH470 pKa = 6.18RR471 pKa = 11.84INWQEE476 pKa = 3.85VARR479 pKa = 11.84LAEE482 pKa = 4.14SLGVAEE488 pKa = 5.35LAPADD493 pKa = 3.98TTEE496 pKa = 3.91GHH498 pKa = 7.34AIAA501 pKa = 5.47
Molecular weight: 55.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13802 |
66 |
716 |
265.4 |
29.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.556 ± 0.777 | 0.935 ± 0.12 |
6.303 ± 0.276 | 7.303 ± 0.283 |
3.485 ± 0.131 | 7.847 ± 0.335 |
2.058 ± 0.198 | 4.847 ± 0.181 |
4.608 ± 0.287 | 7.593 ± 0.365 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.84 ± 0.158 | 3.253 ± 0.168 |
5.086 ± 0.391 | 3.739 ± 0.213 |
7.318 ± 0.353 | 5.688 ± 0.229 |
5.55 ± 0.269 | 6.427 ± 0.265 |
1.746 ± 0.134 | 2.818 ± 0.193 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
