
Haloarcula hispanica pleomorphic virus 1
Taxonomy: Viruses; Monodnaviria; Trapavirae; Saleviricota; Huolimaviricetes; Haloruvirales; Pleolipoviridae; Alphapleolipovirus; Alphapleolipovirus HHPV1
Average proteome isoelectric point is 5.14
Get precalculated fractions of proteins
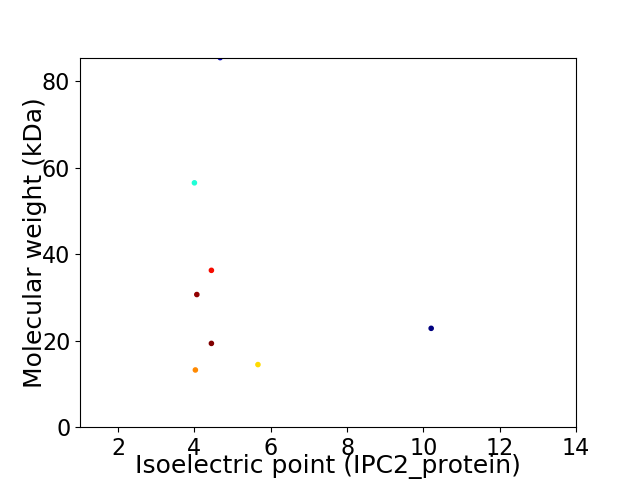
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D3JVC2|D3JVC2_9VIRU ORF5 OS=Haloarcula hispanica pleomorphic virus 1 OX=710112 PE=4 SV=1
MM1 pKa = 7.85SDD3 pKa = 2.84TTMGGSAKK11 pKa = 10.02SSRR14 pKa = 11.84VGRR17 pKa = 11.84RR18 pKa = 11.84TFLKK22 pKa = 9.94TVGVTAGAAAGLHH35 pKa = 5.77HH36 pKa = 7.31TDD38 pKa = 4.06DD39 pKa = 3.87TALDD43 pKa = 3.93PVGEE47 pKa = 4.34ADD49 pKa = 3.31AVVPVVLPGVALAGAAGVGYY69 pKa = 10.48VAGSYY74 pKa = 11.1LEE76 pKa = 5.07DD77 pKa = 3.78NYY79 pKa = 11.35LGDD82 pKa = 3.65SRR84 pKa = 11.84DD85 pKa = 3.52YY86 pKa = 11.3SGYY89 pKa = 9.42TGGDD93 pKa = 3.35ALKK96 pKa = 10.8SAIKK100 pKa = 10.42EE101 pKa = 4.18GATSMKK107 pKa = 10.44VADD110 pKa = 4.56EE111 pKa = 4.45KK112 pKa = 11.68VMSSIQNNIANSQTAGLAKK131 pKa = 10.33GKK133 pKa = 10.24AAVIKK138 pKa = 10.19EE139 pKa = 4.02MNAGNGEE146 pKa = 4.23SAATSAMQSAINEE159 pKa = 4.01YY160 pKa = 10.05FAAIQQNILTHH171 pKa = 5.94YY172 pKa = 10.12NSQLEE177 pKa = 4.36QVKK180 pKa = 9.67HH181 pKa = 6.28HH182 pKa = 6.61VDD184 pKa = 3.34QVANHH189 pKa = 6.57SDD191 pKa = 3.56LTVGDD196 pKa = 3.9VFASEE201 pKa = 4.81NGDD204 pKa = 3.63DD205 pKa = 4.3SPSNMTVNQRR215 pKa = 11.84SLSLVDD221 pKa = 3.65GTDD224 pKa = 3.02ATEE227 pKa = 4.47KK228 pKa = 10.86YY229 pKa = 10.83LIFNTSQDD237 pKa = 2.84GSTYY241 pKa = 10.58EE242 pKa = 4.1SGISLTTATGAADD255 pKa = 3.77FEE257 pKa = 4.75VDD259 pKa = 3.99SFSLSSGAYY268 pKa = 9.71ADD270 pKa = 4.1YY271 pKa = 11.12SDD273 pKa = 3.86EE274 pKa = 4.0FQYY277 pKa = 10.88FQTVPMGNAFDD288 pKa = 4.54AVVTEE293 pKa = 4.1RR294 pKa = 11.84DD295 pKa = 3.48KK296 pKa = 11.79VLDD299 pKa = 4.24DD300 pKa = 4.42LSTFVSDD307 pKa = 4.69VYY309 pKa = 11.0SQYY312 pKa = 11.33SAGDD316 pKa = 3.41IPTEE320 pKa = 4.79DD321 pKa = 3.11IVDD324 pKa = 5.43PITAATEE331 pKa = 3.59LRR333 pKa = 11.84TNYY336 pKa = 10.73DD337 pKa = 3.19GMQGASAHH345 pKa = 6.03AAMLGIPTNADD356 pKa = 2.78QSVYY360 pKa = 7.91MTLEE364 pKa = 4.73DD365 pKa = 4.35DD366 pKa = 5.0GKK368 pKa = 11.12SVWADD373 pKa = 2.94VYY375 pKa = 9.78TNKK378 pKa = 10.65KK379 pKa = 8.04ITDD382 pKa = 3.62SNGNEE387 pKa = 3.69VGFQKK392 pKa = 10.12GTTYY396 pKa = 11.31DD397 pKa = 3.43PTTWSEE403 pKa = 3.73PLYY406 pKa = 10.52IAFEE410 pKa = 4.31YY411 pKa = 10.96TEE413 pKa = 4.38TTDD416 pKa = 3.91SDD418 pKa = 4.54GNVLNSTQQEE428 pKa = 4.7SYY430 pKa = 11.07DD431 pKa = 3.8GEE433 pKa = 4.67TTTTEE438 pKa = 3.65YY439 pKa = 11.31SDD441 pKa = 3.65FVEE444 pKa = 4.53VEE446 pKa = 3.95QPFTIKK452 pKa = 10.54EE453 pKa = 3.88ITVDD457 pKa = 3.88GEE459 pKa = 4.46SKK461 pKa = 10.74QSFKK465 pKa = 8.73TTSRR469 pKa = 11.84NAQTADD475 pKa = 3.13VSKK478 pKa = 10.56LQEE481 pKa = 3.96EE482 pKa = 4.89LEE484 pKa = 4.51QIRR487 pKa = 11.84QTQIDD492 pKa = 4.13MQEE495 pKa = 4.06EE496 pKa = 4.26AQSGGGGGFSVDD508 pKa = 3.59SLSLGGIPGEE518 pKa = 4.2GVLLVGVGIAAWLFGNN534 pKa = 4.69
MM1 pKa = 7.85SDD3 pKa = 2.84TTMGGSAKK11 pKa = 10.02SSRR14 pKa = 11.84VGRR17 pKa = 11.84RR18 pKa = 11.84TFLKK22 pKa = 9.94TVGVTAGAAAGLHH35 pKa = 5.77HH36 pKa = 7.31TDD38 pKa = 4.06DD39 pKa = 3.87TALDD43 pKa = 3.93PVGEE47 pKa = 4.34ADD49 pKa = 3.31AVVPVVLPGVALAGAAGVGYY69 pKa = 10.48VAGSYY74 pKa = 11.1LEE76 pKa = 5.07DD77 pKa = 3.78NYY79 pKa = 11.35LGDD82 pKa = 3.65SRR84 pKa = 11.84DD85 pKa = 3.52YY86 pKa = 11.3SGYY89 pKa = 9.42TGGDD93 pKa = 3.35ALKK96 pKa = 10.8SAIKK100 pKa = 10.42EE101 pKa = 4.18GATSMKK107 pKa = 10.44VADD110 pKa = 4.56EE111 pKa = 4.45KK112 pKa = 11.68VMSSIQNNIANSQTAGLAKK131 pKa = 10.33GKK133 pKa = 10.24AAVIKK138 pKa = 10.19EE139 pKa = 4.02MNAGNGEE146 pKa = 4.23SAATSAMQSAINEE159 pKa = 4.01YY160 pKa = 10.05FAAIQQNILTHH171 pKa = 5.94YY172 pKa = 10.12NSQLEE177 pKa = 4.36QVKK180 pKa = 9.67HH181 pKa = 6.28HH182 pKa = 6.61VDD184 pKa = 3.34QVANHH189 pKa = 6.57SDD191 pKa = 3.56LTVGDD196 pKa = 3.9VFASEE201 pKa = 4.81NGDD204 pKa = 3.63DD205 pKa = 4.3SPSNMTVNQRR215 pKa = 11.84SLSLVDD221 pKa = 3.65GTDD224 pKa = 3.02ATEE227 pKa = 4.47KK228 pKa = 10.86YY229 pKa = 10.83LIFNTSQDD237 pKa = 2.84GSTYY241 pKa = 10.58EE242 pKa = 4.1SGISLTTATGAADD255 pKa = 3.77FEE257 pKa = 4.75VDD259 pKa = 3.99SFSLSSGAYY268 pKa = 9.71ADD270 pKa = 4.1YY271 pKa = 11.12SDD273 pKa = 3.86EE274 pKa = 4.0FQYY277 pKa = 10.88FQTVPMGNAFDD288 pKa = 4.54AVVTEE293 pKa = 4.1RR294 pKa = 11.84DD295 pKa = 3.48KK296 pKa = 11.79VLDD299 pKa = 4.24DD300 pKa = 4.42LSTFVSDD307 pKa = 4.69VYY309 pKa = 11.0SQYY312 pKa = 11.33SAGDD316 pKa = 3.41IPTEE320 pKa = 4.79DD321 pKa = 3.11IVDD324 pKa = 5.43PITAATEE331 pKa = 3.59LRR333 pKa = 11.84TNYY336 pKa = 10.73DD337 pKa = 3.19GMQGASAHH345 pKa = 6.03AAMLGIPTNADD356 pKa = 2.78QSVYY360 pKa = 7.91MTLEE364 pKa = 4.73DD365 pKa = 4.35DD366 pKa = 5.0GKK368 pKa = 11.12SVWADD373 pKa = 2.94VYY375 pKa = 9.78TNKK378 pKa = 10.65KK379 pKa = 8.04ITDD382 pKa = 3.62SNGNEE387 pKa = 3.69VGFQKK392 pKa = 10.12GTTYY396 pKa = 11.31DD397 pKa = 3.43PTTWSEE403 pKa = 3.73PLYY406 pKa = 10.52IAFEE410 pKa = 4.31YY411 pKa = 10.96TEE413 pKa = 4.38TTDD416 pKa = 3.91SDD418 pKa = 4.54GNVLNSTQQEE428 pKa = 4.7SYY430 pKa = 11.07DD431 pKa = 3.8GEE433 pKa = 4.67TTTTEE438 pKa = 3.65YY439 pKa = 11.31SDD441 pKa = 3.65FVEE444 pKa = 4.53VEE446 pKa = 3.95QPFTIKK452 pKa = 10.54EE453 pKa = 3.88ITVDD457 pKa = 3.88GEE459 pKa = 4.46SKK461 pKa = 10.74QSFKK465 pKa = 8.73TTSRR469 pKa = 11.84NAQTADD475 pKa = 3.13VSKK478 pKa = 10.56LQEE481 pKa = 3.96EE482 pKa = 4.89LEE484 pKa = 4.51QIRR487 pKa = 11.84QTQIDD492 pKa = 4.13MQEE495 pKa = 4.06EE496 pKa = 4.26AQSGGGGGFSVDD508 pKa = 3.59SLSLGGIPGEE518 pKa = 4.2GVLLVGVGIAAWLFGNN534 pKa = 4.69
Molecular weight: 56.54 kDa
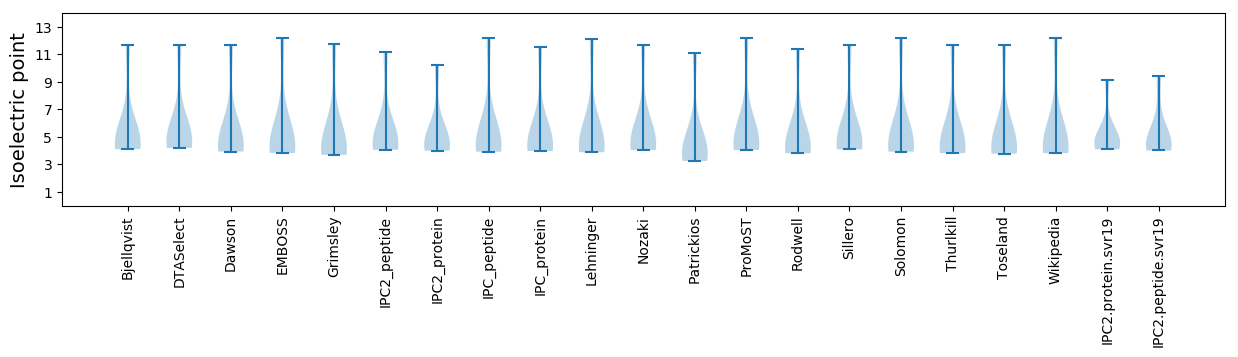
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D3JVC5|D3JVC5_9VIRU ORF8 OS=Haloarcula hispanica pleomorphic virus 1 OX=710112 PE=4 SV=1
MM1 pKa = 7.65SILRR5 pKa = 11.84KK6 pKa = 9.96LIPVGAGGTLVTHH19 pKa = 6.73CASRR23 pKa = 11.84VNIPFVLWWHH33 pKa = 6.65GEE35 pKa = 4.0TGPRR39 pKa = 11.84PVSRR43 pKa = 11.84VDD45 pKa = 3.04ACSLLTTRR53 pKa = 11.84SHH55 pKa = 6.86KK56 pKa = 10.66PNGQYY61 pKa = 11.06NVTGTSRR68 pKa = 11.84RR69 pKa = 11.84AQEE72 pKa = 4.0TPYY75 pKa = 10.77SRR77 pKa = 11.84SQARR81 pKa = 11.84SEE83 pKa = 4.1TGIPRR88 pKa = 11.84AARR91 pKa = 11.84MLARR95 pKa = 11.84PHH97 pKa = 6.37HH98 pKa = 6.08EE99 pKa = 4.15RR100 pKa = 11.84YY101 pKa = 8.28STTSFHH107 pKa = 6.06NQPVAHH113 pKa = 6.43RR114 pKa = 11.84TPPGGPAGGLLSGVLWVGGSLGGYY138 pKa = 8.71PSQARR143 pKa = 11.84TKK145 pKa = 10.33PVAQPNQSSEE155 pKa = 3.6RR156 pKa = 11.84SLFVRR161 pKa = 11.84SRR163 pKa = 11.84TTITQHH169 pKa = 4.57VEE171 pKa = 3.29QRR173 pKa = 11.84RR174 pKa = 11.84PVIIFSEE181 pKa = 4.31QTGSKK186 pKa = 9.8IPLPRR191 pKa = 11.84RR192 pKa = 11.84RR193 pKa = 11.84VIRR196 pKa = 11.84IILRR200 pKa = 11.84CSFFEE205 pKa = 4.15IAA207 pKa = 5.06
MM1 pKa = 7.65SILRR5 pKa = 11.84KK6 pKa = 9.96LIPVGAGGTLVTHH19 pKa = 6.73CASRR23 pKa = 11.84VNIPFVLWWHH33 pKa = 6.65GEE35 pKa = 4.0TGPRR39 pKa = 11.84PVSRR43 pKa = 11.84VDD45 pKa = 3.04ACSLLTTRR53 pKa = 11.84SHH55 pKa = 6.86KK56 pKa = 10.66PNGQYY61 pKa = 11.06NVTGTSRR68 pKa = 11.84RR69 pKa = 11.84AQEE72 pKa = 4.0TPYY75 pKa = 10.77SRR77 pKa = 11.84SQARR81 pKa = 11.84SEE83 pKa = 4.1TGIPRR88 pKa = 11.84AARR91 pKa = 11.84MLARR95 pKa = 11.84PHH97 pKa = 6.37HH98 pKa = 6.08EE99 pKa = 4.15RR100 pKa = 11.84YY101 pKa = 8.28STTSFHH107 pKa = 6.06NQPVAHH113 pKa = 6.43RR114 pKa = 11.84TPPGGPAGGLLSGVLWVGGSLGGYY138 pKa = 8.71PSQARR143 pKa = 11.84TKK145 pKa = 10.33PVAQPNQSSEE155 pKa = 3.6RR156 pKa = 11.84SLFVRR161 pKa = 11.84SRR163 pKa = 11.84TTITQHH169 pKa = 4.57VEE171 pKa = 3.29QRR173 pKa = 11.84RR174 pKa = 11.84PVIIFSEE181 pKa = 4.31QTGSKK186 pKa = 9.8IPLPRR191 pKa = 11.84RR192 pKa = 11.84RR193 pKa = 11.84VIRR196 pKa = 11.84IILRR200 pKa = 11.84CSFFEE205 pKa = 4.15IAA207 pKa = 5.06
Molecular weight: 22.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2571 |
119 |
783 |
321.4 |
34.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.268 ± 0.698 | 1.439 ± 0.625 |
8.518 ± 1.024 | 7.74 ± 0.85 |
2.839 ± 0.663 | 7.74 ± 0.568 |
1.906 ± 0.527 | 3.073 ± 0.694 |
2.995 ± 0.352 | 7.546 ± 1.163 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.867 ± 0.235 | 2.956 ± 0.56 |
4.706 ± 0.74 | 2.8 ± 0.635 |
6.34 ± 1.194 | 7.896 ± 0.678 |
6.884 ± 0.629 | 8.674 ± 0.502 |
1.284 ± 0.195 | 2.528 ± 0.521 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
