
Streptomyces alboflavus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
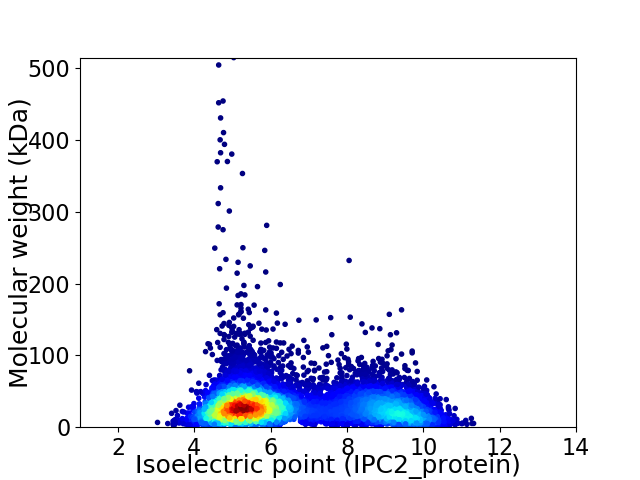
Virtual 2D-PAGE plot for 9243 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
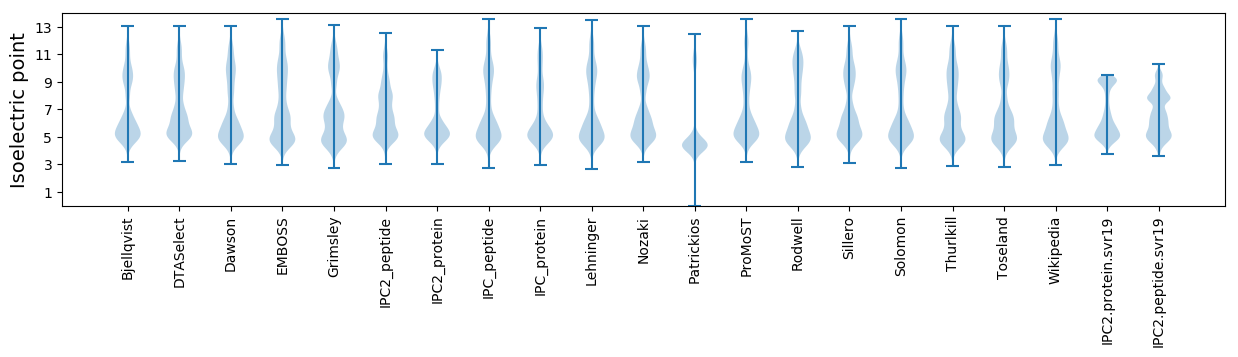
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z1WBM9|A0A1Z1WBM9_9ACTN Uncharacterized protein OS=Streptomyces alboflavus OX=67267 GN=SMD44_03284 PE=4 SV=1
MM1 pKa = 7.92TDD3 pKa = 3.1GQRR6 pKa = 11.84NGAGGWQPHH15 pKa = 5.66PQGEE19 pKa = 4.29YY20 pKa = 10.71DD21 pKa = 3.58ADD23 pKa = 3.31ATAFVQLPEE32 pKa = 4.24GAADD36 pKa = 3.71AVSTPLAAPGHH47 pKa = 6.44GYY49 pKa = 9.98VPPQITVTPTAPDD62 pKa = 3.39ATDD65 pKa = 3.66PAATGAWVLPPEE77 pKa = 4.51VTGVAHH83 pKa = 6.76TGGATQAVSWPDD95 pKa = 3.34PGTQLGTHH103 pKa = 5.83QEE105 pKa = 4.13QQFPAEE111 pKa = 4.08PQVPDD116 pKa = 4.13AYY118 pKa = 10.68DD119 pKa = 3.48PRR121 pKa = 11.84ATGQWTFPDD130 pKa = 3.67AVTPPEE136 pKa = 4.06QNGGQGGVQNGHH148 pKa = 6.71APAAHH153 pKa = 7.08DD154 pKa = 3.96VTGQWSIPVADD165 pKa = 4.18GDD167 pKa = 4.0LPEE170 pKa = 4.58EE171 pKa = 4.29SGEE174 pKa = 4.06YY175 pKa = 9.79SASSLAAQWGGAPPPATLPGGAPAPWATSWPTQAPEE211 pKa = 4.07APQVPQAPQAPQVPQAPEE229 pKa = 4.03LPHH232 pKa = 8.43APEE235 pKa = 5.03APQASPYY242 pKa = 9.66PAAQAFVEE250 pKa = 4.31QAPLGPPAPAQEE262 pKa = 4.41SAPVPPARR270 pKa = 11.84AAEE273 pKa = 4.02EE274 pKa = 4.54TPFEE278 pKa = 4.09RR279 pKa = 11.84TAKK282 pKa = 10.5LFVVKK287 pKa = 9.74QRR289 pKa = 11.84PDD291 pKa = 3.02GSLAPAGSPGDD302 pKa = 3.68VTDD305 pKa = 5.36APDD308 pKa = 3.65APGAPDD314 pKa = 3.71VPGMPDD320 pKa = 2.97ASGEE324 pKa = 4.46TVNPVDD330 pKa = 3.66PAALEE335 pKa = 4.55GDD337 pKa = 4.85FGPQSAASTHH347 pKa = 6.53LAPAADD353 pKa = 3.9PHH355 pKa = 7.3YY356 pKa = 10.44GADD359 pKa = 3.44PAYY362 pKa = 10.57DD363 pKa = 3.44EE364 pKa = 4.32NAYY367 pKa = 10.53DD368 pKa = 3.58EE369 pKa = 4.41SAPYY373 pKa = 10.2EE374 pKa = 4.27EE375 pKa = 4.32NTPYY379 pKa = 11.39DD380 pKa = 3.86MPPVQSADD388 pKa = 3.33LAQSVGLAPAPGFPHH403 pKa = 6.48TVDD406 pKa = 3.05AVYY409 pKa = 9.16PADD412 pKa = 4.15PAHH415 pKa = 6.35ATDD418 pKa = 4.58PSYY421 pKa = 11.72DD422 pKa = 4.08PDD424 pKa = 4.36PALAPPPAHH433 pKa = 6.61PAEE436 pKa = 4.54PAADD440 pKa = 4.0PAHH443 pKa = 6.77PAAVAPAVPPHH454 pKa = 6.33EE455 pKa = 5.33ADD457 pKa = 3.74PAHH460 pKa = 6.37VPAPAQAADD469 pKa = 4.07PAPAGNPAPAPEE481 pKa = 3.93QAHH484 pKa = 5.81AAEE487 pKa = 5.93AIPAADD493 pKa = 4.24PAPAPVPEE501 pKa = 5.33PDD503 pKa = 3.55PAPDD507 pKa = 4.38AGHH510 pKa = 6.36TPDD513 pKa = 4.36PEE515 pKa = 4.07EE516 pKa = 4.16PAAADD521 pKa = 4.08DD522 pKa = 4.9APEE525 pKa = 5.26ADD527 pKa = 5.1DD528 pKa = 4.97PAEE531 pKa = 4.46ADD533 pKa = 3.08QDD535 pKa = 4.19PEE537 pKa = 4.25VEE539 pKa = 4.4HH540 pKa = 7.81APEE543 pKa = 4.8ADD545 pKa = 3.52HH546 pKa = 7.52ASEE549 pKa = 5.4ADD551 pKa = 3.67DD552 pKa = 5.57AEE554 pKa = 5.84ADD556 pKa = 3.94LAADD560 pKa = 4.04SAPVEE565 pKa = 4.12ADD567 pKa = 3.42APVADD572 pKa = 4.97APDD575 pKa = 3.88NAPEE579 pKa = 4.63PDD581 pKa = 3.64APAHH585 pKa = 6.61DD586 pKa = 4.05EE587 pKa = 4.45HH588 pKa = 7.64PLVSYY593 pKa = 8.37VLRR596 pKa = 11.84VNGADD601 pKa = 4.66RR602 pKa = 11.84PVTDD606 pKa = 3.02AWIGEE611 pKa = 4.15SLLYY615 pKa = 10.18VLRR618 pKa = 11.84EE619 pKa = 3.94RR620 pKa = 11.84LGLAGAKK627 pKa = 9.72DD628 pKa = 3.75GCSQGEE634 pKa = 4.12CGACNVQVDD643 pKa = 3.94GRR645 pKa = 11.84LVASCLVPAATAAGSEE661 pKa = 4.08VRR663 pKa = 11.84TVEE666 pKa = 4.04GLAADD671 pKa = 4.46GRR673 pKa = 11.84PSDD676 pKa = 3.9VQRR679 pKa = 11.84ALANCGAVQCGFCVPGMAMTLHH701 pKa = 7.21DD702 pKa = 5.03LLEE705 pKa = 4.8GNPAPSDD712 pKa = 3.69LEE714 pKa = 4.15TRR716 pKa = 11.84RR717 pKa = 11.84ALCGNLCRR725 pKa = 11.84CSGYY729 pKa = 10.39RR730 pKa = 11.84GVLDD734 pKa = 3.66AVRR737 pKa = 11.84DD738 pKa = 4.07VVAEE742 pKa = 4.26RR743 pKa = 11.84EE744 pKa = 4.49AKK746 pKa = 10.02GAAAEE751 pKa = 4.1QRR753 pKa = 11.84DD754 pKa = 4.18DD755 pKa = 4.86ADD757 pKa = 3.46TGTPRR762 pKa = 11.84VPHH765 pKa = 5.37QSGPYY770 pKa = 8.94GQDD773 pKa = 2.85GAQAA777 pKa = 3.71
MM1 pKa = 7.92TDD3 pKa = 3.1GQRR6 pKa = 11.84NGAGGWQPHH15 pKa = 5.66PQGEE19 pKa = 4.29YY20 pKa = 10.71DD21 pKa = 3.58ADD23 pKa = 3.31ATAFVQLPEE32 pKa = 4.24GAADD36 pKa = 3.71AVSTPLAAPGHH47 pKa = 6.44GYY49 pKa = 9.98VPPQITVTPTAPDD62 pKa = 3.39ATDD65 pKa = 3.66PAATGAWVLPPEE77 pKa = 4.51VTGVAHH83 pKa = 6.76TGGATQAVSWPDD95 pKa = 3.34PGTQLGTHH103 pKa = 5.83QEE105 pKa = 4.13QQFPAEE111 pKa = 4.08PQVPDD116 pKa = 4.13AYY118 pKa = 10.68DD119 pKa = 3.48PRR121 pKa = 11.84ATGQWTFPDD130 pKa = 3.67AVTPPEE136 pKa = 4.06QNGGQGGVQNGHH148 pKa = 6.71APAAHH153 pKa = 7.08DD154 pKa = 3.96VTGQWSIPVADD165 pKa = 4.18GDD167 pKa = 4.0LPEE170 pKa = 4.58EE171 pKa = 4.29SGEE174 pKa = 4.06YY175 pKa = 9.79SASSLAAQWGGAPPPATLPGGAPAPWATSWPTQAPEE211 pKa = 4.07APQVPQAPQAPQVPQAPEE229 pKa = 4.03LPHH232 pKa = 8.43APEE235 pKa = 5.03APQASPYY242 pKa = 9.66PAAQAFVEE250 pKa = 4.31QAPLGPPAPAQEE262 pKa = 4.41SAPVPPARR270 pKa = 11.84AAEE273 pKa = 4.02EE274 pKa = 4.54TPFEE278 pKa = 4.09RR279 pKa = 11.84TAKK282 pKa = 10.5LFVVKK287 pKa = 9.74QRR289 pKa = 11.84PDD291 pKa = 3.02GSLAPAGSPGDD302 pKa = 3.68VTDD305 pKa = 5.36APDD308 pKa = 3.65APGAPDD314 pKa = 3.71VPGMPDD320 pKa = 2.97ASGEE324 pKa = 4.46TVNPVDD330 pKa = 3.66PAALEE335 pKa = 4.55GDD337 pKa = 4.85FGPQSAASTHH347 pKa = 6.53LAPAADD353 pKa = 3.9PHH355 pKa = 7.3YY356 pKa = 10.44GADD359 pKa = 3.44PAYY362 pKa = 10.57DD363 pKa = 3.44EE364 pKa = 4.32NAYY367 pKa = 10.53DD368 pKa = 3.58EE369 pKa = 4.41SAPYY373 pKa = 10.2EE374 pKa = 4.27EE375 pKa = 4.32NTPYY379 pKa = 11.39DD380 pKa = 3.86MPPVQSADD388 pKa = 3.33LAQSVGLAPAPGFPHH403 pKa = 6.48TVDD406 pKa = 3.05AVYY409 pKa = 9.16PADD412 pKa = 4.15PAHH415 pKa = 6.35ATDD418 pKa = 4.58PSYY421 pKa = 11.72DD422 pKa = 4.08PDD424 pKa = 4.36PALAPPPAHH433 pKa = 6.61PAEE436 pKa = 4.54PAADD440 pKa = 4.0PAHH443 pKa = 6.77PAAVAPAVPPHH454 pKa = 6.33EE455 pKa = 5.33ADD457 pKa = 3.74PAHH460 pKa = 6.37VPAPAQAADD469 pKa = 4.07PAPAGNPAPAPEE481 pKa = 3.93QAHH484 pKa = 5.81AAEE487 pKa = 5.93AIPAADD493 pKa = 4.24PAPAPVPEE501 pKa = 5.33PDD503 pKa = 3.55PAPDD507 pKa = 4.38AGHH510 pKa = 6.36TPDD513 pKa = 4.36PEE515 pKa = 4.07EE516 pKa = 4.16PAAADD521 pKa = 4.08DD522 pKa = 4.9APEE525 pKa = 5.26ADD527 pKa = 5.1DD528 pKa = 4.97PAEE531 pKa = 4.46ADD533 pKa = 3.08QDD535 pKa = 4.19PEE537 pKa = 4.25VEE539 pKa = 4.4HH540 pKa = 7.81APEE543 pKa = 4.8ADD545 pKa = 3.52HH546 pKa = 7.52ASEE549 pKa = 5.4ADD551 pKa = 3.67DD552 pKa = 5.57AEE554 pKa = 5.84ADD556 pKa = 3.94LAADD560 pKa = 4.04SAPVEE565 pKa = 4.12ADD567 pKa = 3.42APVADD572 pKa = 4.97APDD575 pKa = 3.88NAPEE579 pKa = 4.63PDD581 pKa = 3.64APAHH585 pKa = 6.61DD586 pKa = 4.05EE587 pKa = 4.45HH588 pKa = 7.64PLVSYY593 pKa = 8.37VLRR596 pKa = 11.84VNGADD601 pKa = 4.66RR602 pKa = 11.84PVTDD606 pKa = 3.02AWIGEE611 pKa = 4.15SLLYY615 pKa = 10.18VLRR618 pKa = 11.84EE619 pKa = 3.94RR620 pKa = 11.84LGLAGAKK627 pKa = 9.72DD628 pKa = 3.75GCSQGEE634 pKa = 4.12CGACNVQVDD643 pKa = 3.94GRR645 pKa = 11.84LVASCLVPAATAAGSEE661 pKa = 4.08VRR663 pKa = 11.84TVEE666 pKa = 4.04GLAADD671 pKa = 4.46GRR673 pKa = 11.84PSDD676 pKa = 3.9VQRR679 pKa = 11.84ALANCGAVQCGFCVPGMAMTLHH701 pKa = 7.21DD702 pKa = 5.03LLEE705 pKa = 4.8GNPAPSDD712 pKa = 3.69LEE714 pKa = 4.15TRR716 pKa = 11.84RR717 pKa = 11.84ALCGNLCRR725 pKa = 11.84CSGYY729 pKa = 10.39RR730 pKa = 11.84GVLDD734 pKa = 3.66AVRR737 pKa = 11.84DD738 pKa = 4.07VVAEE742 pKa = 4.26RR743 pKa = 11.84EE744 pKa = 4.49AKK746 pKa = 10.02GAAAEE751 pKa = 4.1QRR753 pKa = 11.84DD754 pKa = 4.18DD755 pKa = 4.86ADD757 pKa = 3.46TGTPRR762 pKa = 11.84VPHH765 pKa = 5.37QSGPYY770 pKa = 8.94GQDD773 pKa = 2.85GAQAA777 pKa = 3.71
Molecular weight: 78.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z1WET9|A0A1Z1WET9_9ACTN RNA polymerase sigma factor SigM OS=Streptomyces alboflavus OX=67267 GN=SMD44_04317 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AVLASRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.74GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AVLASRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.74GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2826290 |
26 |
4855 |
305.8 |
32.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.962 ± 0.039 | 0.832 ± 0.008 |
6.135 ± 0.023 | 5.545 ± 0.024 |
2.652 ± 0.014 | 9.657 ± 0.032 |
2.331 ± 0.013 | 2.895 ± 0.017 |
2.296 ± 0.024 | 10.106 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.702 ± 0.011 | 1.638 ± 0.012 |
6.265 ± 0.027 | 2.682 ± 0.017 |
8.227 ± 0.029 | 5.092 ± 0.02 |
6.085 ± 0.023 | 8.374 ± 0.027 |
1.524 ± 0.011 | 1.999 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
