
Sewage-associated circular DNA virus-18
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.01
Get precalculated fractions of proteins
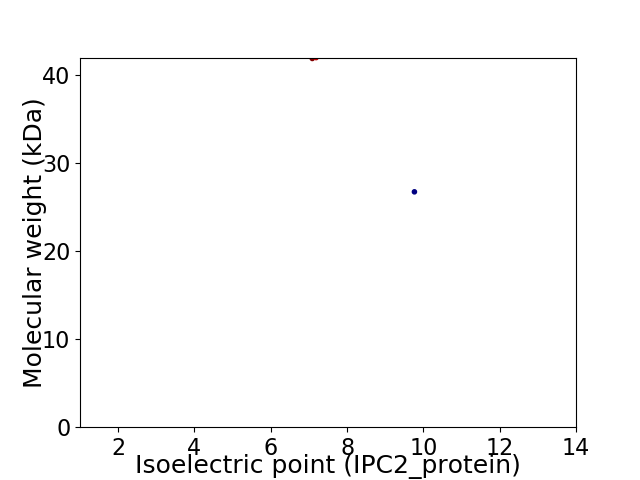
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UH75|A0A0B4UH75_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-18 OX=1592085 PE=3 SV=1
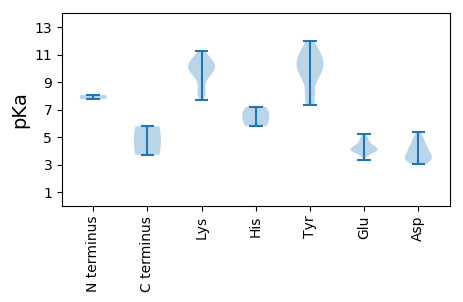
MM1 pKa = 7.76DD2 pKa = 4.92TFPTPNQLILEE13 pKa = 4.41EE14 pKa = 4.95DD15 pKa = 3.44LAEE18 pKa = 4.78PEE20 pKa = 4.14TLEE23 pKa = 4.95APTEE27 pKa = 3.92KK28 pKa = 10.09ATAKK32 pKa = 10.31GFRR35 pKa = 11.84LSGKK39 pKa = 9.4NFTITFPQCDD49 pKa = 3.96VKK51 pKa = 11.25KK52 pKa = 9.68EE53 pKa = 3.9IAVEE57 pKa = 4.2RR58 pKa = 11.84IEE60 pKa = 4.1QKK62 pKa = 10.42FGSEE66 pKa = 3.3IKK68 pKa = 10.57GYY70 pKa = 9.08LVCEE74 pKa = 4.24EE75 pKa = 3.98QHH77 pKa = 7.19KK78 pKa = 11.14DD79 pKa = 3.49GTPHH83 pKa = 5.87LHH85 pKa = 6.43VYY87 pKa = 10.9LSFLNKK93 pKa = 10.06KK94 pKa = 8.48NFKK97 pKa = 9.87CHH99 pKa = 5.79HH100 pKa = 6.53CFNFIGGKK108 pKa = 8.16QGNYY112 pKa = 7.5QVTKK116 pKa = 10.19SVRR119 pKa = 11.84DD120 pKa = 3.32WVTYY124 pKa = 7.31CTKK127 pKa = 10.55GDD129 pKa = 3.51NYY131 pKa = 9.89VAKK134 pKa = 10.31GLDD137 pKa = 3.2VQAIKK142 pKa = 10.79KK143 pKa = 8.07KK144 pKa = 8.09TAPKK148 pKa = 8.27STTVATMLMEE158 pKa = 4.78GKK160 pKa = 9.92SLSEE164 pKa = 4.01INAVDD169 pKa = 3.67PGYY172 pKa = 12.01VMINKK177 pKa = 9.65RR178 pKa = 11.84KK179 pKa = 9.35LEE181 pKa = 4.24EE182 pKa = 3.99YY183 pKa = 9.67EE184 pKa = 3.88SWVTIEE190 pKa = 4.3RR191 pKa = 11.84NKK193 pKa = 10.3KK194 pKa = 10.3SKK196 pKa = 9.15LTWVPPLLDD205 pKa = 4.14GLTDD209 pKa = 3.75ANKK212 pKa = 10.03QICEE216 pKa = 4.51WICSNIRR223 pKa = 11.84QPRR226 pKa = 11.84KK227 pKa = 9.69FKK229 pKa = 10.88APQLFITGPKK239 pKa = 10.04NLGKK243 pKa = 9.26TSLIEE248 pKa = 3.65WLGQYY253 pKa = 10.55LSLYY257 pKa = 9.69HH258 pKa = 6.99IPQTEE263 pKa = 4.07EE264 pKa = 4.55FYY266 pKa = 11.37DD267 pKa = 4.99LYY269 pKa = 10.88TDD271 pKa = 5.35DD272 pKa = 4.22YY273 pKa = 11.01DD274 pKa = 4.94LVVFDD279 pKa = 4.62EE280 pKa = 5.2FKK282 pKa = 10.68GQKK285 pKa = 8.28TIQWMNLFLQGSPMNIRR302 pKa = 11.84KK303 pKa = 9.75KK304 pKa = 10.04GSQYY308 pKa = 10.75MKK310 pKa = 9.84MKK312 pKa = 9.42NLPVIILSNYY322 pKa = 8.2TLGDD326 pKa = 4.39CYY328 pKa = 10.81PKK330 pKa = 10.9ARR332 pKa = 11.84DD333 pKa = 3.66DD334 pKa = 4.05GRR336 pKa = 11.84LEE338 pKa = 3.99TLQARR343 pKa = 11.84LDD345 pKa = 4.02VIEE348 pKa = 4.44VDD350 pKa = 4.5SFIDD354 pKa = 4.32FYY356 pKa = 11.53KK357 pKa = 10.82DD358 pKa = 3.01RR359 pKa = 11.84SDD361 pKa = 3.42ILL363 pKa = 3.7
MM1 pKa = 7.76DD2 pKa = 4.92TFPTPNQLILEE13 pKa = 4.41EE14 pKa = 4.95DD15 pKa = 3.44LAEE18 pKa = 4.78PEE20 pKa = 4.14TLEE23 pKa = 4.95APTEE27 pKa = 3.92KK28 pKa = 10.09ATAKK32 pKa = 10.31GFRR35 pKa = 11.84LSGKK39 pKa = 9.4NFTITFPQCDD49 pKa = 3.96VKK51 pKa = 11.25KK52 pKa = 9.68EE53 pKa = 3.9IAVEE57 pKa = 4.2RR58 pKa = 11.84IEE60 pKa = 4.1QKK62 pKa = 10.42FGSEE66 pKa = 3.3IKK68 pKa = 10.57GYY70 pKa = 9.08LVCEE74 pKa = 4.24EE75 pKa = 3.98QHH77 pKa = 7.19KK78 pKa = 11.14DD79 pKa = 3.49GTPHH83 pKa = 5.87LHH85 pKa = 6.43VYY87 pKa = 10.9LSFLNKK93 pKa = 10.06KK94 pKa = 8.48NFKK97 pKa = 9.87CHH99 pKa = 5.79HH100 pKa = 6.53CFNFIGGKK108 pKa = 8.16QGNYY112 pKa = 7.5QVTKK116 pKa = 10.19SVRR119 pKa = 11.84DD120 pKa = 3.32WVTYY124 pKa = 7.31CTKK127 pKa = 10.55GDD129 pKa = 3.51NYY131 pKa = 9.89VAKK134 pKa = 10.31GLDD137 pKa = 3.2VQAIKK142 pKa = 10.79KK143 pKa = 8.07KK144 pKa = 8.09TAPKK148 pKa = 8.27STTVATMLMEE158 pKa = 4.78GKK160 pKa = 9.92SLSEE164 pKa = 4.01INAVDD169 pKa = 3.67PGYY172 pKa = 12.01VMINKK177 pKa = 9.65RR178 pKa = 11.84KK179 pKa = 9.35LEE181 pKa = 4.24EE182 pKa = 3.99YY183 pKa = 9.67EE184 pKa = 3.88SWVTIEE190 pKa = 4.3RR191 pKa = 11.84NKK193 pKa = 10.3KK194 pKa = 10.3SKK196 pKa = 9.15LTWVPPLLDD205 pKa = 4.14GLTDD209 pKa = 3.75ANKK212 pKa = 10.03QICEE216 pKa = 4.51WICSNIRR223 pKa = 11.84QPRR226 pKa = 11.84KK227 pKa = 9.69FKK229 pKa = 10.88APQLFITGPKK239 pKa = 10.04NLGKK243 pKa = 9.26TSLIEE248 pKa = 3.65WLGQYY253 pKa = 10.55LSLYY257 pKa = 9.69HH258 pKa = 6.99IPQTEE263 pKa = 4.07EE264 pKa = 4.55FYY266 pKa = 11.37DD267 pKa = 4.99LYY269 pKa = 10.88TDD271 pKa = 5.35DD272 pKa = 4.22YY273 pKa = 11.01DD274 pKa = 4.94LVVFDD279 pKa = 4.62EE280 pKa = 5.2FKK282 pKa = 10.68GQKK285 pKa = 8.28TIQWMNLFLQGSPMNIRR302 pKa = 11.84KK303 pKa = 9.75KK304 pKa = 10.04GSQYY308 pKa = 10.75MKK310 pKa = 9.84MKK312 pKa = 9.42NLPVIILSNYY322 pKa = 8.2TLGDD326 pKa = 4.39CYY328 pKa = 10.81PKK330 pKa = 10.9ARR332 pKa = 11.84DD333 pKa = 3.66DD334 pKa = 4.05GRR336 pKa = 11.84LEE338 pKa = 3.99TLQARR343 pKa = 11.84LDD345 pKa = 4.02VIEE348 pKa = 4.44VDD350 pKa = 4.5SFIDD354 pKa = 4.32FYY356 pKa = 11.53KK357 pKa = 10.82DD358 pKa = 3.01RR359 pKa = 11.84SDD361 pKa = 3.42ILL363 pKa = 3.7
Molecular weight: 41.87 kDa
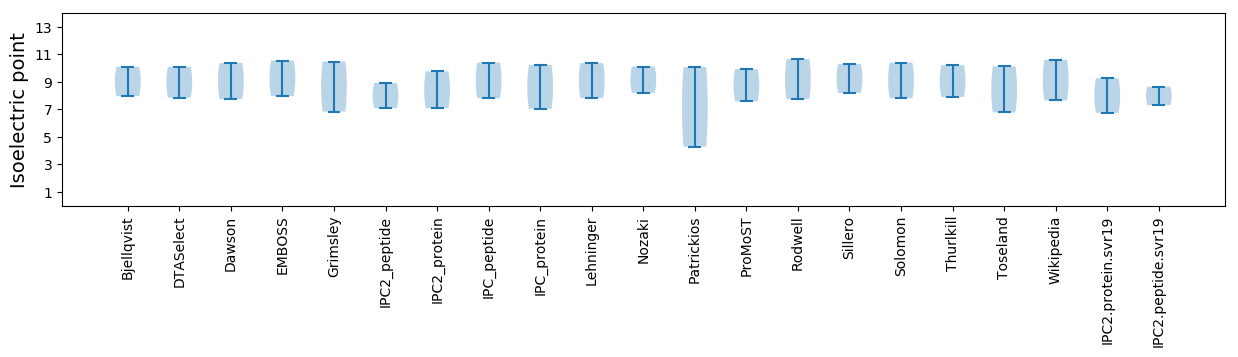
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UH75|A0A0B4UH75_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-18 OX=1592085 PE=3 SV=1
MM1 pKa = 8.05RR2 pKa = 11.84GPTKK6 pKa = 10.39RR7 pKa = 11.84NYY9 pKa = 9.68KK10 pKa = 9.98GSSRR14 pKa = 11.84KK15 pKa = 8.98IAAARR20 pKa = 11.84AAVGRR25 pKa = 11.84ANAVLASRR33 pKa = 11.84SSMSSGSYY41 pKa = 10.28GPPASRR47 pKa = 11.84GFYY50 pKa = 9.85GQYY53 pKa = 10.08SLRR56 pKa = 11.84GRR58 pKa = 11.84AEE60 pKa = 4.04LKK62 pKa = 10.67FVDD65 pKa = 3.91ATSTNAAVTTTWQGALINGIAQGADD90 pKa = 3.13FNQRR94 pKa = 11.84IGRR97 pKa = 11.84KK98 pKa = 8.46AQMKK102 pKa = 9.86SVLFNGNFFPGTTAAEE118 pKa = 4.08NASQGVYY125 pKa = 9.97LRR127 pKa = 11.84VVIVYY132 pKa = 9.65DD133 pKa = 3.55SQPNSGTFPGGTDD146 pKa = 3.53FLGANDD152 pKa = 4.78PNTPLNLNNRR162 pKa = 11.84DD163 pKa = 3.55RR164 pKa = 11.84FSILIDD170 pKa = 3.2VRR172 pKa = 11.84KK173 pKa = 10.22QIGSYY178 pKa = 10.32LFNGTPALTAGSPQNAYY195 pKa = 7.38WNKK198 pKa = 9.58YY199 pKa = 8.24KK200 pKa = 10.58KK201 pKa = 10.16CNKK204 pKa = 7.73EE205 pKa = 4.27TIFSGTAATLGSISTGAMYY224 pKa = 10.42IFFVGDD230 pKa = 3.38FNGVGMIDD238 pKa = 4.22FYY240 pKa = 11.41TRR242 pKa = 11.84VRR244 pKa = 11.84YY245 pKa = 8.36TDD247 pKa = 3.09MM248 pKa = 5.78
MM1 pKa = 8.05RR2 pKa = 11.84GPTKK6 pKa = 10.39RR7 pKa = 11.84NYY9 pKa = 9.68KK10 pKa = 9.98GSSRR14 pKa = 11.84KK15 pKa = 8.98IAAARR20 pKa = 11.84AAVGRR25 pKa = 11.84ANAVLASRR33 pKa = 11.84SSMSSGSYY41 pKa = 10.28GPPASRR47 pKa = 11.84GFYY50 pKa = 9.85GQYY53 pKa = 10.08SLRR56 pKa = 11.84GRR58 pKa = 11.84AEE60 pKa = 4.04LKK62 pKa = 10.67FVDD65 pKa = 3.91ATSTNAAVTTTWQGALINGIAQGADD90 pKa = 3.13FNQRR94 pKa = 11.84IGRR97 pKa = 11.84KK98 pKa = 8.46AQMKK102 pKa = 9.86SVLFNGNFFPGTTAAEE118 pKa = 4.08NASQGVYY125 pKa = 9.97LRR127 pKa = 11.84VVIVYY132 pKa = 9.65DD133 pKa = 3.55SQPNSGTFPGGTDD146 pKa = 3.53FLGANDD152 pKa = 4.78PNTPLNLNNRR162 pKa = 11.84DD163 pKa = 3.55RR164 pKa = 11.84FSILIDD170 pKa = 3.2VRR172 pKa = 11.84KK173 pKa = 10.22QIGSYY178 pKa = 10.32LFNGTPALTAGSPQNAYY195 pKa = 7.38WNKK198 pKa = 9.58YY199 pKa = 8.24KK200 pKa = 10.58KK201 pKa = 10.16CNKK204 pKa = 7.73EE205 pKa = 4.27TIFSGTAATLGSISTGAMYY224 pKa = 10.42IFFVGDD230 pKa = 3.38FNGVGMIDD238 pKa = 4.22FYY240 pKa = 11.41TRR242 pKa = 11.84VRR244 pKa = 11.84YY245 pKa = 8.36TDD247 pKa = 3.09MM248 pKa = 5.78
Molecular weight: 26.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
611 |
248 |
363 |
305.5 |
34.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.71 ± 2.373 | 1.473 ± 0.608 |
5.401 ± 0.777 | 4.583 ± 1.916 |
5.237 ± 0.461 | 8.02 ± 1.858 |
0.982 ± 0.558 | 5.728 ± 0.505 |
8.02 ± 2.036 | 7.692 ± 1.392 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.291 ± 0.073 | 5.892 ± 1.005 |
4.419 ± 0.22 | 4.255 ± 0.356 |
4.746 ± 1.198 | 6.219 ± 1.277 |
7.201 ± 0.261 | 5.237 ± 0.003 |
1.309 ± 0.286 | 4.583 ± 0.145 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
