
Podoviridae sp. ctviO18
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; unclassified Podoviridae
Average proteome isoelectric point is 7.08
Get precalculated fractions of proteins
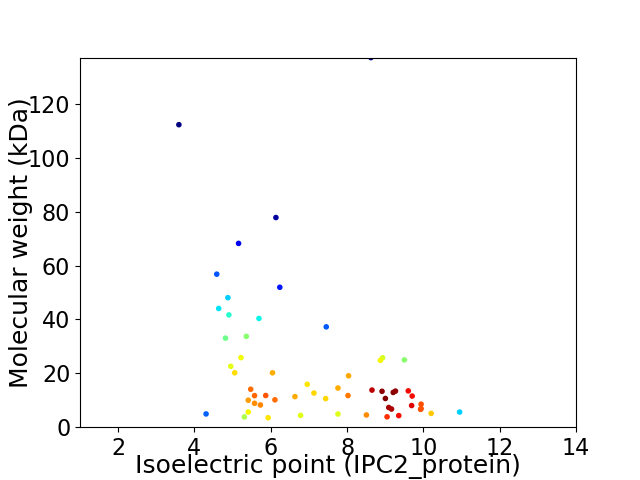
Virtual 2D-PAGE plot for 58 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q2W4G5|A0A5Q2W4G5_9CAUD Uncharacterized protein OS=Podoviridae sp. ctviO18 OX=2656716 PE=4 SV=1
MM1 pKa = 7.4MKK3 pKa = 9.76MKK5 pKa = 10.54NRR7 pKa = 11.84TFLIFPIISVGLIVIALLGIQIVYY31 pKa = 10.81APVQTVKK38 pKa = 10.95NLTAGDD44 pKa = 4.1TIGTLPAIIASGSRR58 pKa = 11.84NLLVPRR64 pKa = 11.84DD65 pKa = 3.6RR66 pKa = 11.84DD67 pKa = 3.21ADD69 pKa = 3.97FVFGGQSSVSAQGKK83 pKa = 8.84FVLDD87 pKa = 4.01DD88 pKa = 3.62VGSTMNLEE96 pKa = 4.28LDD98 pKa = 4.85GIASISKK105 pKa = 10.19FSLIDD110 pKa = 3.34SSTGITMVDD119 pKa = 3.54CDD121 pKa = 4.88GDD123 pKa = 3.96AQALGWDD130 pKa = 3.75LTNQRR135 pKa = 11.84FFCGDD140 pKa = 3.63DD141 pKa = 3.48DD142 pKa = 4.83SGGGGGGTLIEE153 pKa = 4.32VQNSFVDD160 pKa = 3.84LGSFSSISFDD170 pKa = 3.3AGHH173 pKa = 6.1FTATDD178 pKa = 3.5TTGEE182 pKa = 4.04ATIKK186 pKa = 10.52LDD188 pKa = 3.22WGAGGPASLSQAEE201 pKa = 4.73TITGNWVNTDD211 pKa = 3.76NPWDD215 pKa = 3.53ISTEE219 pKa = 4.21TNLTAGTDD227 pKa = 3.62LTLTNDD233 pKa = 4.18DD234 pKa = 4.18LTLDD238 pKa = 3.63STLTQNFTFNAAGTAVIVTNNARR261 pKa = 11.84FGTASVSTGFEE272 pKa = 4.49AITYY276 pKa = 9.84ASASLFQGSAFAGIPGAEE294 pKa = 4.5CSDD297 pKa = 4.6AGDD300 pKa = 3.61TLAWANGQFTCGSDD314 pKa = 3.12ASGSGGASVEE324 pKa = 4.26FQEE327 pKa = 5.18SGTDD331 pKa = 3.04KK332 pKa = 11.35GSFSSVSFDD341 pKa = 2.95AGKK344 pKa = 10.77FNITDD349 pKa = 3.41TTGEE353 pKa = 3.88VDD355 pKa = 5.49IDD357 pKa = 3.78IDD359 pKa = 3.72WTSGGGPASKK369 pKa = 10.25SQANTWSQLQTFTLGASSTLNFEE392 pKa = 4.36AVGYY396 pKa = 10.49ASASALLGSAFTSVGDD412 pKa = 4.26CNDD415 pKa = 3.08ATEE418 pKa = 5.02AIGWTTTGIFNCRR431 pKa = 11.84SVQDD435 pKa = 4.02LDD437 pKa = 3.76STLTALAAYY446 pKa = 7.33NTNGLLTQTAADD458 pKa = 3.96TFTGRR463 pKa = 11.84TITGTSNQITVANGDD478 pKa = 3.98GVSGNPTLSIPSIFIAPGTASVTTDD503 pKa = 3.61FEE505 pKa = 6.01AIGFASASAFRR516 pKa = 11.84GSAFSGGDD524 pKa = 3.58CDD526 pKa = 5.48GATQKK531 pKa = 9.47LTWDD535 pKa = 3.65TTNGTFGCATDD546 pKa = 4.51DD547 pKa = 4.13DD548 pKa = 4.58VPEE551 pKa = 4.43VGDD554 pKa = 4.29FGALVGGLAIDD565 pKa = 4.2NNSGTLDD572 pKa = 4.21LDD574 pKa = 3.42PTEE577 pKa = 5.32LINNRR582 pKa = 11.84TWSDD586 pKa = 3.44GTSDD590 pKa = 5.0SSIVWTWNLSAGTDD604 pKa = 3.51PTLTFGDD611 pKa = 4.39GVITFGQGATISTNFEE627 pKa = 4.16VKK629 pKa = 9.08GTASISAFTLPDD641 pKa = 3.51KK642 pKa = 11.36DD643 pKa = 3.63GGTLGDD649 pKa = 4.65CEE651 pKa = 4.55ASTEE655 pKa = 4.14KK656 pKa = 10.71VVYY659 pKa = 10.59DD660 pKa = 4.12LATKK664 pKa = 10.61KK665 pKa = 10.27FDD667 pKa = 4.21CGTDD671 pKa = 3.36QSGSGGSGGNVSDD684 pKa = 4.89GLDD687 pKa = 3.12IANSGGTYY695 pKa = 10.31LAIASLSFDD704 pKa = 3.21ASHH707 pKa = 6.48FTFTNTASDD716 pKa = 4.26GYY718 pKa = 10.67IRR720 pKa = 11.84LDD722 pKa = 3.31WGSGGPASLSEE733 pKa = 4.49AEE735 pKa = 4.47TISGNWVNTASPWADD750 pKa = 3.06NEE752 pKa = 4.39VVDD755 pKa = 5.09TISIIGGIIGANSISGTQTTTGTLTIGDD783 pKa = 4.29NGDD786 pKa = 4.05SIIIDD791 pKa = 3.52ASNWDD796 pKa = 3.51VSTLGKK802 pKa = 10.5ADD804 pKa = 3.93FLSASVSTNFEE815 pKa = 3.79ATGYY819 pKa = 10.88ASASKK824 pKa = 10.88YY825 pKa = 10.43FGAGLGSMSGTDD837 pKa = 3.46GCSAAGDD844 pKa = 3.78TLNYY848 pKa = 8.98TFSTGVFSCGSDD860 pKa = 2.91ASGGGGVSSNSLNFDD875 pKa = 3.43EE876 pKa = 5.08FQNPLVLDD884 pKa = 3.63VMASVQFGVFNWEE897 pKa = 3.47FDD899 pKa = 4.09LNSTGDD905 pKa = 4.48FIISDD910 pKa = 3.89NNSNLVTFWDD920 pKa = 3.45TGGASFSTPVTGTIDD935 pKa = 3.34EE936 pKa = 4.99LLTFGATSAIPPSTSFATFDD956 pKa = 3.34TRR958 pKa = 11.84NDD960 pKa = 3.32FTVLDD965 pKa = 4.28FGDD968 pKa = 3.76GAVASKK974 pKa = 10.82SIFVGVMPRR983 pKa = 11.84DD984 pKa = 3.69YY985 pKa = 11.49DD986 pKa = 4.12DD987 pKa = 3.44GTMTIVIHH995 pKa = 6.23FAATSATSGNAVWDD1009 pKa = 3.84VEE1011 pKa = 4.0FDD1013 pKa = 3.8RR1014 pKa = 11.84VGTTLDD1020 pKa = 3.31TDD1022 pKa = 3.89SASWATARR1030 pKa = 11.84TATCAVSGTAGRR1042 pKa = 11.84LSTCLITFTKK1052 pKa = 10.37AQADD1056 pKa = 3.99GVLKK1060 pKa = 11.12GEE1062 pKa = 4.66LFRR1065 pKa = 11.84MRR1067 pKa = 11.84VSRR1070 pKa = 11.84DD1071 pKa = 3.12TTDD1074 pKa = 3.35AADD1077 pKa = 3.74TVTGTDD1083 pKa = 3.22LEE1085 pKa = 4.5LYY1087 pKa = 9.89YY1088 pKa = 10.11IEE1090 pKa = 5.8IIQQ1093 pKa = 4.02
MM1 pKa = 7.4MKK3 pKa = 9.76MKK5 pKa = 10.54NRR7 pKa = 11.84TFLIFPIISVGLIVIALLGIQIVYY31 pKa = 10.81APVQTVKK38 pKa = 10.95NLTAGDD44 pKa = 4.1TIGTLPAIIASGSRR58 pKa = 11.84NLLVPRR64 pKa = 11.84DD65 pKa = 3.6RR66 pKa = 11.84DD67 pKa = 3.21ADD69 pKa = 3.97FVFGGQSSVSAQGKK83 pKa = 8.84FVLDD87 pKa = 4.01DD88 pKa = 3.62VGSTMNLEE96 pKa = 4.28LDD98 pKa = 4.85GIASISKK105 pKa = 10.19FSLIDD110 pKa = 3.34SSTGITMVDD119 pKa = 3.54CDD121 pKa = 4.88GDD123 pKa = 3.96AQALGWDD130 pKa = 3.75LTNQRR135 pKa = 11.84FFCGDD140 pKa = 3.63DD141 pKa = 3.48DD142 pKa = 4.83SGGGGGGTLIEE153 pKa = 4.32VQNSFVDD160 pKa = 3.84LGSFSSISFDD170 pKa = 3.3AGHH173 pKa = 6.1FTATDD178 pKa = 3.5TTGEE182 pKa = 4.04ATIKK186 pKa = 10.52LDD188 pKa = 3.22WGAGGPASLSQAEE201 pKa = 4.73TITGNWVNTDD211 pKa = 3.76NPWDD215 pKa = 3.53ISTEE219 pKa = 4.21TNLTAGTDD227 pKa = 3.62LTLTNDD233 pKa = 4.18DD234 pKa = 4.18LTLDD238 pKa = 3.63STLTQNFTFNAAGTAVIVTNNARR261 pKa = 11.84FGTASVSTGFEE272 pKa = 4.49AITYY276 pKa = 9.84ASASLFQGSAFAGIPGAEE294 pKa = 4.5CSDD297 pKa = 4.6AGDD300 pKa = 3.61TLAWANGQFTCGSDD314 pKa = 3.12ASGSGGASVEE324 pKa = 4.26FQEE327 pKa = 5.18SGTDD331 pKa = 3.04KK332 pKa = 11.35GSFSSVSFDD341 pKa = 2.95AGKK344 pKa = 10.77FNITDD349 pKa = 3.41TTGEE353 pKa = 3.88VDD355 pKa = 5.49IDD357 pKa = 3.78IDD359 pKa = 3.72WTSGGGPASKK369 pKa = 10.25SQANTWSQLQTFTLGASSTLNFEE392 pKa = 4.36AVGYY396 pKa = 10.49ASASALLGSAFTSVGDD412 pKa = 4.26CNDD415 pKa = 3.08ATEE418 pKa = 5.02AIGWTTTGIFNCRR431 pKa = 11.84SVQDD435 pKa = 4.02LDD437 pKa = 3.76STLTALAAYY446 pKa = 7.33NTNGLLTQTAADD458 pKa = 3.96TFTGRR463 pKa = 11.84TITGTSNQITVANGDD478 pKa = 3.98GVSGNPTLSIPSIFIAPGTASVTTDD503 pKa = 3.61FEE505 pKa = 6.01AIGFASASAFRR516 pKa = 11.84GSAFSGGDD524 pKa = 3.58CDD526 pKa = 5.48GATQKK531 pKa = 9.47LTWDD535 pKa = 3.65TTNGTFGCATDD546 pKa = 4.51DD547 pKa = 4.13DD548 pKa = 4.58VPEE551 pKa = 4.43VGDD554 pKa = 4.29FGALVGGLAIDD565 pKa = 4.2NNSGTLDD572 pKa = 4.21LDD574 pKa = 3.42PTEE577 pKa = 5.32LINNRR582 pKa = 11.84TWSDD586 pKa = 3.44GTSDD590 pKa = 5.0SSIVWTWNLSAGTDD604 pKa = 3.51PTLTFGDD611 pKa = 4.39GVITFGQGATISTNFEE627 pKa = 4.16VKK629 pKa = 9.08GTASISAFTLPDD641 pKa = 3.51KK642 pKa = 11.36DD643 pKa = 3.63GGTLGDD649 pKa = 4.65CEE651 pKa = 4.55ASTEE655 pKa = 4.14KK656 pKa = 10.71VVYY659 pKa = 10.59DD660 pKa = 4.12LATKK664 pKa = 10.61KK665 pKa = 10.27FDD667 pKa = 4.21CGTDD671 pKa = 3.36QSGSGGSGGNVSDD684 pKa = 4.89GLDD687 pKa = 3.12IANSGGTYY695 pKa = 10.31LAIASLSFDD704 pKa = 3.21ASHH707 pKa = 6.48FTFTNTASDD716 pKa = 4.26GYY718 pKa = 10.67IRR720 pKa = 11.84LDD722 pKa = 3.31WGSGGPASLSEE733 pKa = 4.49AEE735 pKa = 4.47TISGNWVNTASPWADD750 pKa = 3.06NEE752 pKa = 4.39VVDD755 pKa = 5.09TISIIGGIIGANSISGTQTTTGTLTIGDD783 pKa = 4.29NGDD786 pKa = 4.05SIIIDD791 pKa = 3.52ASNWDD796 pKa = 3.51VSTLGKK802 pKa = 10.5ADD804 pKa = 3.93FLSASVSTNFEE815 pKa = 3.79ATGYY819 pKa = 10.88ASASKK824 pKa = 10.88YY825 pKa = 10.43FGAGLGSMSGTDD837 pKa = 3.46GCSAAGDD844 pKa = 3.78TLNYY848 pKa = 8.98TFSTGVFSCGSDD860 pKa = 2.91ASGGGGVSSNSLNFDD875 pKa = 3.43EE876 pKa = 5.08FQNPLVLDD884 pKa = 3.63VMASVQFGVFNWEE897 pKa = 3.47FDD899 pKa = 4.09LNSTGDD905 pKa = 4.48FIISDD910 pKa = 3.89NNSNLVTFWDD920 pKa = 3.45TGGASFSTPVTGTIDD935 pKa = 3.34EE936 pKa = 4.99LLTFGATSAIPPSTSFATFDD956 pKa = 3.34TRR958 pKa = 11.84NDD960 pKa = 3.32FTVLDD965 pKa = 4.28FGDD968 pKa = 3.76GAVASKK974 pKa = 10.82SIFVGVMPRR983 pKa = 11.84DD984 pKa = 3.69YY985 pKa = 11.49DD986 pKa = 4.12DD987 pKa = 3.44GTMTIVIHH995 pKa = 6.23FAATSATSGNAVWDD1009 pKa = 3.84VEE1011 pKa = 4.0FDD1013 pKa = 3.8RR1014 pKa = 11.84VGTTLDD1020 pKa = 3.31TDD1022 pKa = 3.89SASWATARR1030 pKa = 11.84TATCAVSGTAGRR1042 pKa = 11.84LSTCLITFTKK1052 pKa = 10.37AQADD1056 pKa = 3.99GVLKK1060 pKa = 11.12GEE1062 pKa = 4.66LFRR1065 pKa = 11.84MRR1067 pKa = 11.84VSRR1070 pKa = 11.84DD1071 pKa = 3.12TTDD1074 pKa = 3.35AADD1077 pKa = 3.74TVTGTDD1083 pKa = 3.22LEE1085 pKa = 4.5LYY1087 pKa = 9.89YY1088 pKa = 10.11IEE1090 pKa = 5.8IIQQ1093 pKa = 4.02
Molecular weight: 112.3 kDa
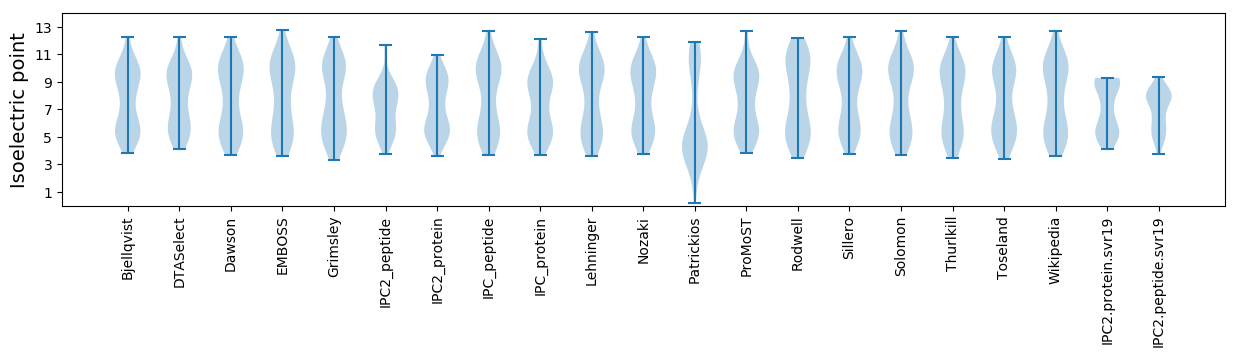
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q2WAV3|A0A5Q2WAV3_9CAUD Uncharacterized protein OS=Podoviridae sp. ctviO18 OX=2656716 PE=4 SV=1
MM1 pKa = 7.31PQAFEE6 pKa = 3.74QCRR9 pKa = 11.84ANGGKK14 pKa = 9.24IRR16 pKa = 11.84TKK18 pKa = 9.47TLPGGKK24 pKa = 8.98YY25 pKa = 7.11VHH27 pKa = 6.79FCILNGKK34 pKa = 7.69SYY36 pKa = 10.96RR37 pKa = 11.84GYY39 pKa = 9.14TKK41 pKa = 9.09TARR44 pKa = 11.84TSRR47 pKa = 11.84YY48 pKa = 8.61KK49 pKa = 10.87GRR51 pKa = 11.84SSEE54 pKa = 4.33SKK56 pKa = 10.97SNNQYY61 pKa = 11.3
MM1 pKa = 7.31PQAFEE6 pKa = 3.74QCRR9 pKa = 11.84ANGGKK14 pKa = 9.24IRR16 pKa = 11.84TKK18 pKa = 9.47TLPGGKK24 pKa = 8.98YY25 pKa = 7.11VHH27 pKa = 6.79FCILNGKK34 pKa = 7.69SYY36 pKa = 10.96RR37 pKa = 11.84GYY39 pKa = 9.14TKK41 pKa = 9.09TARR44 pKa = 11.84TSRR47 pKa = 11.84YY48 pKa = 8.61KK49 pKa = 10.87GRR51 pKa = 11.84SSEE54 pKa = 4.33SKK56 pKa = 10.97SNNQYY61 pKa = 11.3
Molecular weight: 6.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11718 |
30 |
1238 |
202.0 |
22.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.075 ± 0.418 | 1.135 ± 0.191 |
6.298 ± 0.381 | 6.281 ± 0.559 |
3.951 ± 0.241 | 8.679 ± 0.752 |
1.434 ± 0.196 | 6.494 ± 0.241 |
7.937 ± 0.799 | 7.518 ± 0.213 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.005 ± 0.198 | 5.069 ± 0.227 |
3.849 ± 0.29 | 3.354 ± 0.216 |
4.642 ± 0.395 | 6.93 ± 0.518 |
6.4 ± 0.649 | 5.863 ± 0.256 |
1.63 ± 0.185 | 3.456 ± 0.367 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
