
Caenispirillum salinarum AK4
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillaceae; Caenispirillum; Caenispirillum salinarum
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
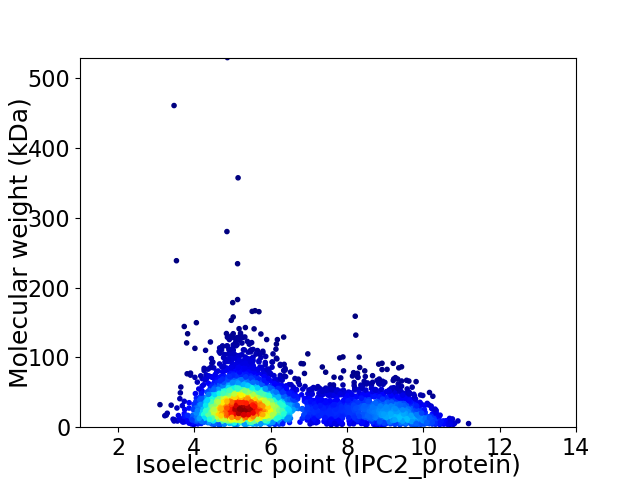
Virtual 2D-PAGE plot for 4574 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K9GRX7|K9GRX7_9PROT Alpha-1 4 glucan phosphorylase OS=Caenispirillum salinarum AK4 OX=1238182 GN=C882_0927 PE=3 SV=1
MM1 pKa = 7.49KK2 pKa = 10.22KK3 pKa = 10.34VLLGTSALIAAAAISAPAMAQEE25 pKa = 4.21QIQLGIGGKK34 pKa = 7.61MEE36 pKa = 4.03QYY38 pKa = 10.72FGVTSIDD45 pKa = 3.43DD46 pKa = 3.85DD47 pKa = 4.12AAGFDD52 pKa = 3.94PTTTGINTDD61 pKa = 3.17AEE63 pKa = 5.03VYY65 pKa = 8.13FTGATTLDD73 pKa = 3.25NGLTVGAVIQLEE85 pKa = 4.33AQTNANVTFNADD97 pKa = 2.86EE98 pKa = 3.99QYY100 pKa = 11.54AYY102 pKa = 9.92IEE104 pKa = 4.36GAFGQLRR111 pKa = 11.84AGQKK115 pKa = 10.4NGILQDD121 pKa = 4.03LAHH124 pKa = 6.91TAPQIGISFLDD135 pKa = 3.6AASFVPSAGLAVGTSQSKK153 pKa = 10.49LVDD156 pKa = 3.47TTVSDD161 pKa = 4.34DD162 pKa = 3.82SASVSYY168 pKa = 8.87ITPTLAGFSAGVTFNPNPGGALQANQEE195 pKa = 4.47DD196 pKa = 4.82GAGTHH201 pKa = 6.33NKK203 pKa = 9.14FQVGLAYY210 pKa = 10.57NGEE213 pKa = 4.06FSGVGIGVDD222 pKa = 3.55GSYY225 pKa = 8.18QTEE228 pKa = 4.11EE229 pKa = 4.17ADD231 pKa = 3.93DD232 pKa = 5.41AIVGADD238 pKa = 4.59DD239 pKa = 3.59PTVWRR244 pKa = 11.84AGLNLAYY251 pKa = 10.21AGFQAGGSYY260 pKa = 10.31LRR262 pKa = 11.84SEE264 pKa = 4.39DD265 pKa = 3.72TADD268 pKa = 3.67TEE270 pKa = 4.6TTTWDD275 pKa = 2.94AGVAYY280 pKa = 9.84GVGPYY285 pKa = 9.45QVGVGYY291 pKa = 10.47VQTEE295 pKa = 4.01VDD297 pKa = 4.71DD298 pKa = 4.2GTVANDD304 pKa = 2.75VDD306 pKa = 4.59EE307 pKa = 4.31YY308 pKa = 11.33RR309 pKa = 11.84QVQLSGSYY317 pKa = 10.03QMGPGIKK324 pKa = 10.02AVGGLFWYY332 pKa = 10.0EE333 pKa = 3.98SEE335 pKa = 4.69SNVGLGTTTTNEE347 pKa = 3.71RR348 pKa = 11.84DD349 pKa = 3.41GGGGIVGLALTFF361 pKa = 4.17
MM1 pKa = 7.49KK2 pKa = 10.22KK3 pKa = 10.34VLLGTSALIAAAAISAPAMAQEE25 pKa = 4.21QIQLGIGGKK34 pKa = 7.61MEE36 pKa = 4.03QYY38 pKa = 10.72FGVTSIDD45 pKa = 3.43DD46 pKa = 3.85DD47 pKa = 4.12AAGFDD52 pKa = 3.94PTTTGINTDD61 pKa = 3.17AEE63 pKa = 5.03VYY65 pKa = 8.13FTGATTLDD73 pKa = 3.25NGLTVGAVIQLEE85 pKa = 4.33AQTNANVTFNADD97 pKa = 2.86EE98 pKa = 3.99QYY100 pKa = 11.54AYY102 pKa = 9.92IEE104 pKa = 4.36GAFGQLRR111 pKa = 11.84AGQKK115 pKa = 10.4NGILQDD121 pKa = 4.03LAHH124 pKa = 6.91TAPQIGISFLDD135 pKa = 3.6AASFVPSAGLAVGTSQSKK153 pKa = 10.49LVDD156 pKa = 3.47TTVSDD161 pKa = 4.34DD162 pKa = 3.82SASVSYY168 pKa = 8.87ITPTLAGFSAGVTFNPNPGGALQANQEE195 pKa = 4.47DD196 pKa = 4.82GAGTHH201 pKa = 6.33NKK203 pKa = 9.14FQVGLAYY210 pKa = 10.57NGEE213 pKa = 4.06FSGVGIGVDD222 pKa = 3.55GSYY225 pKa = 8.18QTEE228 pKa = 4.11EE229 pKa = 4.17ADD231 pKa = 3.93DD232 pKa = 5.41AIVGADD238 pKa = 4.59DD239 pKa = 3.59PTVWRR244 pKa = 11.84AGLNLAYY251 pKa = 10.21AGFQAGGSYY260 pKa = 10.31LRR262 pKa = 11.84SEE264 pKa = 4.39DD265 pKa = 3.72TADD268 pKa = 3.67TEE270 pKa = 4.6TTTWDD275 pKa = 2.94AGVAYY280 pKa = 9.84GVGPYY285 pKa = 9.45QVGVGYY291 pKa = 10.47VQTEE295 pKa = 4.01VDD297 pKa = 4.71DD298 pKa = 4.2GTVANDD304 pKa = 2.75VDD306 pKa = 4.59EE307 pKa = 4.31YY308 pKa = 11.33RR309 pKa = 11.84QVQLSGSYY317 pKa = 10.03QMGPGIKK324 pKa = 10.02AVGGLFWYY332 pKa = 10.0EE333 pKa = 3.98SEE335 pKa = 4.69SNVGLGTTTTNEE347 pKa = 3.71RR348 pKa = 11.84DD349 pKa = 3.41GGGGIVGLALTFF361 pKa = 4.17
Molecular weight: 37.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K9GT92|K9GT92_9PROT Tryptophan-rich sensory protein OS=Caenispirillum salinarum AK4 OX=1238182 GN=C882_0981 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 8.99RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.7GFRR19 pKa = 11.84SRR21 pKa = 11.84SATVGGRR28 pKa = 11.84RR29 pKa = 11.84VLANRR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.48GRR39 pKa = 11.84KK40 pKa = 8.72RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 8.99RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.7GFRR19 pKa = 11.84SRR21 pKa = 11.84SATVGGRR28 pKa = 11.84RR29 pKa = 11.84VLANRR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.48GRR39 pKa = 11.84KK40 pKa = 8.72RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1438899 |
37 |
5007 |
314.6 |
33.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.233 ± 0.067 | 0.875 ± 0.011 |
6.299 ± 0.036 | 6.064 ± 0.036 |
3.28 ± 0.023 | 8.844 ± 0.036 |
2.145 ± 0.019 | 4.211 ± 0.025 |
2.807 ± 0.03 | 10.069 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.635 ± 0.018 | 2.183 ± 0.022 |
5.652 ± 0.032 | 2.807 ± 0.022 |
7.514 ± 0.042 | 4.454 ± 0.023 |
5.537 ± 0.034 | 7.992 ± 0.033 |
1.363 ± 0.017 | 2.035 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
