
Akanthomyces lecanii RCEF 1005
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Hypocreomycetidae; Hypocreales; Cordycipitaceae; Akanthomyces; Akanthomyces lecanii
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
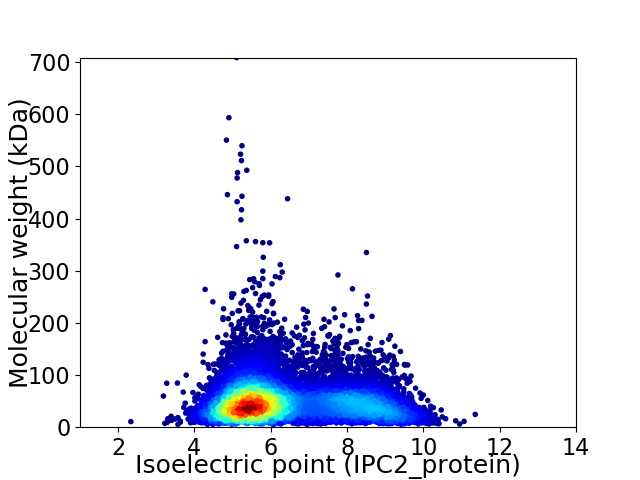
Virtual 2D-PAGE plot for 11019 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A168H3Q9|A0A168H3Q9_CORDF Uncharacterized protein OS=Akanthomyces lecanii RCEF 1005 OX=1081108 GN=LEL_04094 PE=4 SV=1
MM1 pKa = 7.46FLQTILAATAFGALASAAPTQSRR24 pKa = 11.84STGGKK29 pKa = 9.24LAVYY33 pKa = 9.01WGAEE37 pKa = 4.13DD38 pKa = 5.16SSTTLDD44 pKa = 4.07DD45 pKa = 3.66VCGDD49 pKa = 3.4SSYY52 pKa = 11.43DD53 pKa = 3.44IVNLAFLSYY62 pKa = 9.85FFKK65 pKa = 10.99DD66 pKa = 2.77GGYY69 pKa = 9.51PEE71 pKa = 5.71LSISTLGGPSDD82 pKa = 3.79AQQAAGATSLQDD94 pKa = 3.36GSEE97 pKa = 4.23LVPAIQKK104 pKa = 8.28CQKK107 pKa = 9.17NGKK110 pKa = 9.24RR111 pKa = 11.84VILSMGGAQGYY122 pKa = 11.19ADD124 pKa = 3.64VTLQSDD130 pKa = 3.66AQGQQIAQTIWDD142 pKa = 4.43LFLGGTNNPSLRR154 pKa = 11.84PFGDD158 pKa = 3.28VKK160 pKa = 11.22LDD162 pKa = 4.15GVDD165 pKa = 4.26LDD167 pKa = 4.54NEE169 pKa = 4.72SGNPTGYY176 pKa = 11.11VAMTKK181 pKa = 7.8QFRR184 pKa = 11.84SLMNSDD190 pKa = 3.38SSKK193 pKa = 10.95QYY195 pKa = 11.27VLTAAPQCPFPDD207 pKa = 3.44ASEE210 pKa = 4.26PLDD213 pKa = 3.71VVQLLDD219 pKa = 3.79YY220 pKa = 11.14VWVQFYY226 pKa = 11.57NNGDD230 pKa = 3.84CNIAQSGFNDD240 pKa = 4.13AVRR243 pKa = 11.84TWSQGIGNATLFIGALASGADD264 pKa = 3.63GDD266 pKa = 3.91QGYY269 pKa = 10.73VDD271 pKa = 4.81ASTFTSALQGVEE283 pKa = 4.45AMNLPNYY290 pKa = 8.91GGAMLWEE297 pKa = 4.42AQLAVNNGNYY307 pKa = 7.98QQQIRR312 pKa = 11.84GSLL315 pKa = 3.3
MM1 pKa = 7.46FLQTILAATAFGALASAAPTQSRR24 pKa = 11.84STGGKK29 pKa = 9.24LAVYY33 pKa = 9.01WGAEE37 pKa = 4.13DD38 pKa = 5.16SSTTLDD44 pKa = 4.07DD45 pKa = 3.66VCGDD49 pKa = 3.4SSYY52 pKa = 11.43DD53 pKa = 3.44IVNLAFLSYY62 pKa = 9.85FFKK65 pKa = 10.99DD66 pKa = 2.77GGYY69 pKa = 9.51PEE71 pKa = 5.71LSISTLGGPSDD82 pKa = 3.79AQQAAGATSLQDD94 pKa = 3.36GSEE97 pKa = 4.23LVPAIQKK104 pKa = 8.28CQKK107 pKa = 9.17NGKK110 pKa = 9.24RR111 pKa = 11.84VILSMGGAQGYY122 pKa = 11.19ADD124 pKa = 3.64VTLQSDD130 pKa = 3.66AQGQQIAQTIWDD142 pKa = 4.43LFLGGTNNPSLRR154 pKa = 11.84PFGDD158 pKa = 3.28VKK160 pKa = 11.22LDD162 pKa = 4.15GVDD165 pKa = 4.26LDD167 pKa = 4.54NEE169 pKa = 4.72SGNPTGYY176 pKa = 11.11VAMTKK181 pKa = 7.8QFRR184 pKa = 11.84SLMNSDD190 pKa = 3.38SSKK193 pKa = 10.95QYY195 pKa = 11.27VLTAAPQCPFPDD207 pKa = 3.44ASEE210 pKa = 4.26PLDD213 pKa = 3.71VVQLLDD219 pKa = 3.79YY220 pKa = 11.14VWVQFYY226 pKa = 11.57NNGDD230 pKa = 3.84CNIAQSGFNDD240 pKa = 4.13AVRR243 pKa = 11.84TWSQGIGNATLFIGALASGADD264 pKa = 3.63GDD266 pKa = 3.91QGYY269 pKa = 10.73VDD271 pKa = 4.81ASTFTSALQGVEE283 pKa = 4.45AMNLPNYY290 pKa = 8.91GGAMLWEE297 pKa = 4.42AQLAVNNGNYY307 pKa = 7.98QQQIRR312 pKa = 11.84GSLL315 pKa = 3.3
Molecular weight: 33.25 kDa
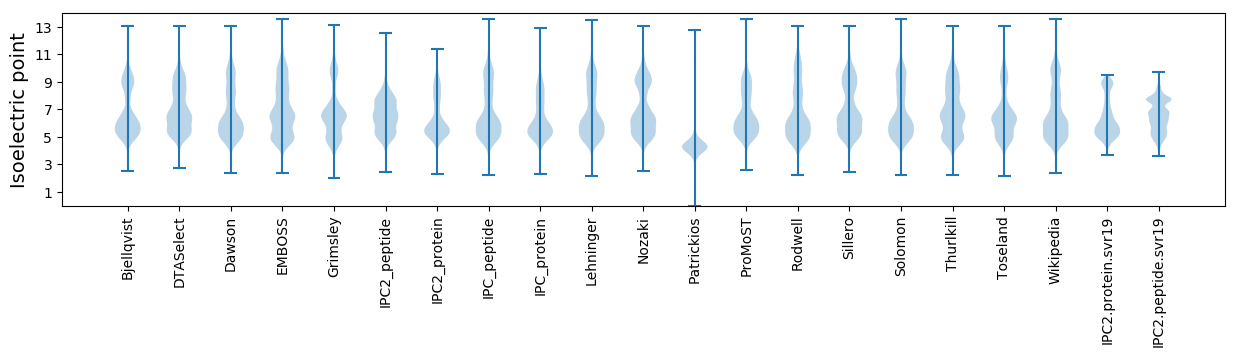
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A168CSM9|A0A168CSM9_CORDF Ferredoxin reductase-type FAD-binding domain protein OS=Akanthomyces lecanii RCEF 1005 OX=1081108 GN=LEL_08976 PE=4 SV=1
MM1 pKa = 7.1QPAKK5 pKa = 10.22RR6 pKa = 11.84GRR8 pKa = 11.84SAKK11 pKa = 10.03RR12 pKa = 11.84GQPGKK17 pKa = 10.2RR18 pKa = 11.84GQSGKK23 pKa = 10.22RR24 pKa = 11.84GQPGKK29 pKa = 10.31RR30 pKa = 11.84GQPGKK35 pKa = 10.31RR36 pKa = 11.84GQPGKK41 pKa = 10.31RR42 pKa = 11.84GQPGKK47 pKa = 10.31RR48 pKa = 11.84GQPGKK53 pKa = 10.31RR54 pKa = 11.84GQPGKK59 pKa = 10.31RR60 pKa = 11.84GQPGKK65 pKa = 10.31RR66 pKa = 11.84GQPGKK71 pKa = 10.31RR72 pKa = 11.84GQPGKK77 pKa = 10.31RR78 pKa = 11.84GQPGKK83 pKa = 10.31RR84 pKa = 11.84GQPGKK89 pKa = 10.31RR90 pKa = 11.84GQPGKK95 pKa = 10.31RR96 pKa = 11.84GQPGKK101 pKa = 10.31RR102 pKa = 11.84GQPGKK107 pKa = 10.31RR108 pKa = 11.84GQPGKK113 pKa = 10.31RR114 pKa = 11.84GQPGKK119 pKa = 10.31RR120 pKa = 11.84GQPGKK125 pKa = 10.31RR126 pKa = 11.84GQPGKK131 pKa = 10.31RR132 pKa = 11.84GQPGKK137 pKa = 10.31RR138 pKa = 11.84GQPGKK143 pKa = 10.31RR144 pKa = 11.84GQPGKK149 pKa = 10.31RR150 pKa = 11.84GQPGKK155 pKa = 10.31RR156 pKa = 11.84GQPGKK161 pKa = 10.31RR162 pKa = 11.84GQPGKK167 pKa = 10.31RR168 pKa = 11.84GQPGKK173 pKa = 10.31RR174 pKa = 11.84GQPGKK179 pKa = 10.31RR180 pKa = 11.84GQPGKK185 pKa = 10.31RR186 pKa = 11.84GQPGKK191 pKa = 10.27RR192 pKa = 11.84GRR194 pKa = 11.84PGKK197 pKa = 10.16RR198 pKa = 11.84GQPGKK203 pKa = 10.31RR204 pKa = 11.84GQPGKK209 pKa = 10.32RR210 pKa = 11.84GQPTEE215 pKa = 4.15EE216 pKa = 4.15NSTGQLSRR224 pKa = 11.84RR225 pKa = 11.84QWAATVIRR233 pKa = 4.59
MM1 pKa = 7.1QPAKK5 pKa = 10.22RR6 pKa = 11.84GRR8 pKa = 11.84SAKK11 pKa = 10.03RR12 pKa = 11.84GQPGKK17 pKa = 10.2RR18 pKa = 11.84GQSGKK23 pKa = 10.22RR24 pKa = 11.84GQPGKK29 pKa = 10.31RR30 pKa = 11.84GQPGKK35 pKa = 10.31RR36 pKa = 11.84GQPGKK41 pKa = 10.31RR42 pKa = 11.84GQPGKK47 pKa = 10.31RR48 pKa = 11.84GQPGKK53 pKa = 10.31RR54 pKa = 11.84GQPGKK59 pKa = 10.31RR60 pKa = 11.84GQPGKK65 pKa = 10.31RR66 pKa = 11.84GQPGKK71 pKa = 10.31RR72 pKa = 11.84GQPGKK77 pKa = 10.31RR78 pKa = 11.84GQPGKK83 pKa = 10.31RR84 pKa = 11.84GQPGKK89 pKa = 10.31RR90 pKa = 11.84GQPGKK95 pKa = 10.31RR96 pKa = 11.84GQPGKK101 pKa = 10.31RR102 pKa = 11.84GQPGKK107 pKa = 10.31RR108 pKa = 11.84GQPGKK113 pKa = 10.31RR114 pKa = 11.84GQPGKK119 pKa = 10.31RR120 pKa = 11.84GQPGKK125 pKa = 10.31RR126 pKa = 11.84GQPGKK131 pKa = 10.31RR132 pKa = 11.84GQPGKK137 pKa = 10.31RR138 pKa = 11.84GQPGKK143 pKa = 10.31RR144 pKa = 11.84GQPGKK149 pKa = 10.31RR150 pKa = 11.84GQPGKK155 pKa = 10.31RR156 pKa = 11.84GQPGKK161 pKa = 10.31RR162 pKa = 11.84GQPGKK167 pKa = 10.31RR168 pKa = 11.84GQPGKK173 pKa = 10.31RR174 pKa = 11.84GQPGKK179 pKa = 10.31RR180 pKa = 11.84GQPGKK185 pKa = 10.31RR186 pKa = 11.84GQPGKK191 pKa = 10.27RR192 pKa = 11.84GRR194 pKa = 11.84PGKK197 pKa = 10.16RR198 pKa = 11.84GQPGKK203 pKa = 10.31RR204 pKa = 11.84GQPGKK209 pKa = 10.32RR210 pKa = 11.84GQPTEE215 pKa = 4.15EE216 pKa = 4.15NSTGQLSRR224 pKa = 11.84RR225 pKa = 11.84QWAATVIRR233 pKa = 4.59
Molecular weight: 24.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5350041 |
49 |
6480 |
485.5 |
53.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.71 ± 0.025 | 1.241 ± 0.008 |
5.891 ± 0.017 | 5.876 ± 0.026 |
3.674 ± 0.013 | 7.101 ± 0.022 |
2.378 ± 0.01 | 4.632 ± 0.017 |
4.715 ± 0.021 | 8.826 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.296 ± 0.01 | 3.508 ± 0.013 |
5.81 ± 0.023 | 4.029 ± 0.016 |
6.053 ± 0.02 | 7.96 ± 0.026 |
5.899 ± 0.017 | 6.252 ± 0.016 |
1.461 ± 0.009 | 2.687 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
