
Pan troglodytes polyomavirus 4
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Alphapolyomavirus
Average proteome isoelectric point is 7.1
Get precalculated fractions of proteins
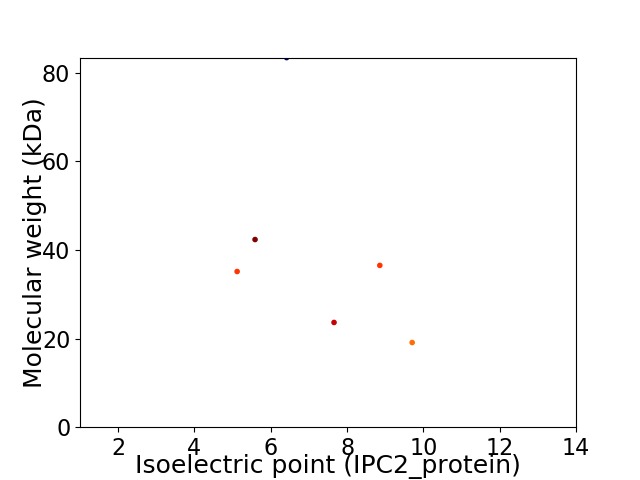
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K7QKH3|K7QKH3_9POLY Minor capsid protein OS=Pan troglodytes polyomavirus 4 OX=1891736 PE=3 SV=1
MM1 pKa = 7.47GGVLSIIVDD10 pKa = 4.37IIEE13 pKa = 4.02LTAEE17 pKa = 3.97VTAATGFSLEE27 pKa = 4.31AIINGEE33 pKa = 3.67ALAAVEE39 pKa = 4.4AQLSSLATVEE49 pKa = 4.17GLSGVEE55 pKa = 4.03ALASLGITTEE65 pKa = 4.02QYY67 pKa = 11.46SFLTSVPGMLSNAVGLGVVFQTVSGASALVSAGIATSYY105 pKa = 10.59AKK107 pKa = 10.13EE108 pKa = 3.83VSVVNRR114 pKa = 11.84NMALVPWRR122 pKa = 11.84DD123 pKa = 2.93PDD125 pKa = 3.7YY126 pKa = 11.54YY127 pKa = 11.05DD128 pKa = 3.18ILFPGVQSFSYY139 pKa = 10.2AINVVADD146 pKa = 3.62WSGSLFHH153 pKa = 7.57AIGRR157 pKa = 11.84YY158 pKa = 8.79IWDD161 pKa = 3.83TVTSEE166 pKa = 3.58ARR168 pKa = 11.84RR169 pKa = 11.84QIGHH173 pKa = 7.72ASNQVIVRR181 pKa = 11.84GTHH184 pKa = 6.53AFQDD188 pKa = 3.55SLARR192 pKa = 11.84ILEE195 pKa = 3.92NARR198 pKa = 11.84WVVQAGPTNIYY209 pKa = 10.79NFLDD213 pKa = 3.23NYY215 pKa = 10.15YY216 pKa = 10.67RR217 pKa = 11.84EE218 pKa = 4.71LPPVNPPQARR228 pKa = 11.84QMYY231 pKa = 9.35RR232 pKa = 11.84RR233 pKa = 11.84LGQKK237 pKa = 9.79YY238 pKa = 7.99RR239 pKa = 11.84FARR242 pKa = 11.84EE243 pKa = 3.82AYY245 pKa = 9.82QIPDD249 pKa = 3.79RR250 pKa = 11.84YY251 pKa = 10.88NLANSEE257 pKa = 4.38TEE259 pKa = 3.74SGEE262 pKa = 3.98TVEE265 pKa = 5.56FYY267 pKa = 10.85KK268 pKa = 10.99HH269 pKa = 6.39PGGANQRR276 pKa = 11.84VTPDD280 pKa = 2.51WMLPLILGLYY290 pKa = 10.53GDD292 pKa = 4.64ITPTWSTYY300 pKa = 6.34VHH302 pKa = 6.85EE303 pKa = 4.89IEE305 pKa = 5.82EE306 pKa = 4.25EE307 pKa = 4.12DD308 pKa = 3.49EE309 pKa = 4.32SQKK312 pKa = 10.88KK313 pKa = 9.04KK314 pKa = 9.72RR315 pKa = 11.84RR316 pKa = 11.84RR317 pKa = 11.84YY318 pKa = 9.21
MM1 pKa = 7.47GGVLSIIVDD10 pKa = 4.37IIEE13 pKa = 4.02LTAEE17 pKa = 3.97VTAATGFSLEE27 pKa = 4.31AIINGEE33 pKa = 3.67ALAAVEE39 pKa = 4.4AQLSSLATVEE49 pKa = 4.17GLSGVEE55 pKa = 4.03ALASLGITTEE65 pKa = 4.02QYY67 pKa = 11.46SFLTSVPGMLSNAVGLGVVFQTVSGASALVSAGIATSYY105 pKa = 10.59AKK107 pKa = 10.13EE108 pKa = 3.83VSVVNRR114 pKa = 11.84NMALVPWRR122 pKa = 11.84DD123 pKa = 2.93PDD125 pKa = 3.7YY126 pKa = 11.54YY127 pKa = 11.05DD128 pKa = 3.18ILFPGVQSFSYY139 pKa = 10.2AINVVADD146 pKa = 3.62WSGSLFHH153 pKa = 7.57AIGRR157 pKa = 11.84YY158 pKa = 8.79IWDD161 pKa = 3.83TVTSEE166 pKa = 3.58ARR168 pKa = 11.84RR169 pKa = 11.84QIGHH173 pKa = 7.72ASNQVIVRR181 pKa = 11.84GTHH184 pKa = 6.53AFQDD188 pKa = 3.55SLARR192 pKa = 11.84ILEE195 pKa = 3.92NARR198 pKa = 11.84WVVQAGPTNIYY209 pKa = 10.79NFLDD213 pKa = 3.23NYY215 pKa = 10.15YY216 pKa = 10.67RR217 pKa = 11.84EE218 pKa = 4.71LPPVNPPQARR228 pKa = 11.84QMYY231 pKa = 9.35RR232 pKa = 11.84RR233 pKa = 11.84LGQKK237 pKa = 9.79YY238 pKa = 7.99RR239 pKa = 11.84FARR242 pKa = 11.84EE243 pKa = 3.82AYY245 pKa = 9.82QIPDD249 pKa = 3.79RR250 pKa = 11.84YY251 pKa = 10.88NLANSEE257 pKa = 4.38TEE259 pKa = 3.74SGEE262 pKa = 3.98TVEE265 pKa = 5.56FYY267 pKa = 10.85KK268 pKa = 10.99HH269 pKa = 6.39PGGANQRR276 pKa = 11.84VTPDD280 pKa = 2.51WMLPLILGLYY290 pKa = 10.53GDD292 pKa = 4.64ITPTWSTYY300 pKa = 6.34VHH302 pKa = 6.85EE303 pKa = 4.89IEE305 pKa = 5.82EE306 pKa = 4.25EE307 pKa = 4.12DD308 pKa = 3.49EE309 pKa = 4.32SQKK312 pKa = 10.88KK313 pKa = 9.04KK314 pKa = 9.72RR315 pKa = 11.84RR316 pKa = 11.84RR317 pKa = 11.84YY318 pKa = 9.21
Molecular weight: 35.15 kDa
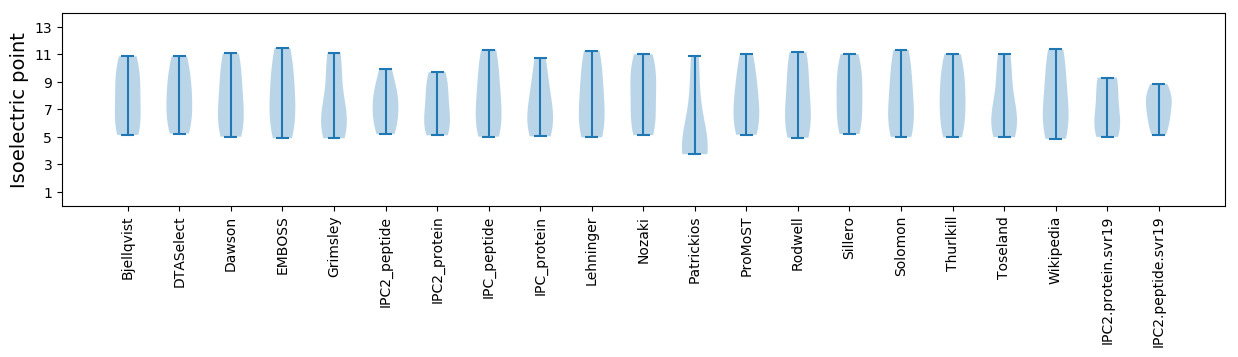
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5J6XVL7|A0A5J6XVL7_9POLY Middle T antigen OS=Pan troglodytes polyomavirus 4 OX=1891736 PE=4 SV=1
MM1 pKa = 7.72GPRR4 pKa = 11.84NLRR7 pKa = 11.84SGGMHH12 pKa = 6.62NFHH15 pKa = 7.16PVHH18 pKa = 7.02PSTSAPQLTTDD29 pKa = 3.64EE30 pKa = 5.03EE31 pKa = 4.33EE32 pKa = 4.4DD33 pKa = 3.15PTYY36 pKa = 11.05IVMKK40 pKa = 10.28SLNPVMKK47 pKa = 9.97KK48 pKa = 10.44DD49 pKa = 3.67PQPAPKK55 pKa = 9.45PVSSDD60 pKa = 3.26EE61 pKa = 4.2EE62 pKa = 4.34RR63 pKa = 11.84PPTPPPRR70 pKa = 11.84RR71 pKa = 11.84IFSPLLPRR79 pKa = 11.84PRR81 pKa = 11.84RR82 pKa = 11.84VPLLHH87 pKa = 6.4MPEE90 pKa = 4.3IPGGTLPARR99 pKa = 11.84LTSPLTSLHH108 pKa = 6.49RR109 pKa = 11.84PPNVLPLNLLEE120 pKa = 6.15KK121 pKa = 10.31EE122 pKa = 4.08IQTLRR127 pKa = 11.84RR128 pKa = 11.84QVSLAHH134 pKa = 7.16RR135 pKa = 11.84RR136 pKa = 11.84RR137 pKa = 11.84QRR139 pKa = 11.84DD140 pKa = 3.25LKK142 pKa = 10.88FIMILSIFQILFLIILVMLYY162 pKa = 9.98IVIKK166 pKa = 10.36HH167 pKa = 5.54
MM1 pKa = 7.72GPRR4 pKa = 11.84NLRR7 pKa = 11.84SGGMHH12 pKa = 6.62NFHH15 pKa = 7.16PVHH18 pKa = 7.02PSTSAPQLTTDD29 pKa = 3.64EE30 pKa = 5.03EE31 pKa = 4.33EE32 pKa = 4.4DD33 pKa = 3.15PTYY36 pKa = 11.05IVMKK40 pKa = 10.28SLNPVMKK47 pKa = 9.97KK48 pKa = 10.44DD49 pKa = 3.67PQPAPKK55 pKa = 9.45PVSSDD60 pKa = 3.26EE61 pKa = 4.2EE62 pKa = 4.34RR63 pKa = 11.84PPTPPPRR70 pKa = 11.84RR71 pKa = 11.84IFSPLLPRR79 pKa = 11.84PRR81 pKa = 11.84RR82 pKa = 11.84VPLLHH87 pKa = 6.4MPEE90 pKa = 4.3IPGGTLPARR99 pKa = 11.84LTSPLTSLHH108 pKa = 6.49RR109 pKa = 11.84PPNVLPLNLLEE120 pKa = 6.15KK121 pKa = 10.31EE122 pKa = 4.08IQTLRR127 pKa = 11.84RR128 pKa = 11.84QVSLAHH134 pKa = 7.16RR135 pKa = 11.84RR136 pKa = 11.84RR137 pKa = 11.84QRR139 pKa = 11.84DD140 pKa = 3.25LKK142 pKa = 10.88FIMILSIFQILFLIILVMLYY162 pKa = 9.98IVIKK166 pKa = 10.36HH167 pKa = 5.54
Molecular weight: 19.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2108 |
167 |
730 |
351.3 |
40.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.602 ± 1.019 | 2.324 ± 0.608 |
5.455 ± 0.518 | 5.93 ± 0.359 |
4.317 ± 0.505 | 5.218 ± 0.77 |
2.277 ± 0.414 | 5.171 ± 0.384 |
6.072 ± 0.972 | 10.769 ± 0.904 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.226 ± 0.605 | 4.886 ± 0.473 |
7.258 ± 1.058 | 4.696 ± 0.274 |
5.977 ± 1.008 | 7.163 ± 0.671 |
4.934 ± 0.843 | 5.455 ± 0.85 |
1.091 ± 0.287 | 3.178 ± 0.409 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
